Gene
KWMTBOMO16867 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011844
Annotation
protein_disulfide_isomerase_precursor_[Bombyx_mori]
Full name
Protein disulfide-isomerase
Location in the cell
ER Reliability : 2.628
Sequence
CDS
ATGGAGATCTTGGAGTTCTTCGGCATGAAGAAGGATGAGGTTCCATCTGCCCGTCTGATCGCCCTTGAACAAGACATGGCCAAATACAAGCCCAGCAGTAATGAGCTGTCGCCTAACGCTATTGAGGAATTCGTTCAATCGTTCTTCGACGGCACCCTGAAGCAGCATCTGCTGAGCGAGGACCTGCCCTCGGACTGGGCCGCCAAACCCGTCAAAGTGCTCGTCGCCGCCAACTTCGACGAAGTCGTCTTCGACACAACTAAGAAGGTCCTCGTCGAATTCTACGCTCCTTGGTGCGGCCACTGCAAACAGCTGGTCCCTATCTACGACAAGCTCGGCGAGCACTTTGAGAACGACGATGACGTCATCATTGCCAAAATCGACGCCACCGCCAACGAACTGGAACACACCAAGATCACGTCCTTCCCAACGATCAAACTTTACAGCAAGGACAACCAGGTCCACGACTACAACGGCGAGAGGACACTGGCCGGCCTCACCAAGTTCGTTGAGACCGACGGTGAAGGCGCCGAGCCAGGACCTTCTGTGACCGAATTCGAAGAGGAGGAAGATGAATTCGTTCAATCGTTCTTCGACGGCACCCTGAAGCAGCATCTGCTGAGCGAGGACCTGCCCTCGGACTGGGCCGCCAAACCCGTCAAAGTGCTCGTCGCCGCCAACTTCGACGAAGTCGTCTTCGACACAACTAAGAAGGTCCTCGTCGAATTCTACGCTCCTTGGTAA
Protein
MEILEFFGMKKDEVPSARLIALEQDMAKYKPSSNELSPNAIEEFVQSFFDGTLKQHLLSEDLPSDWAAKPVKVLVAANFDEVVFDTTKKVLVEFYAPWCGHCKQLVPIYDKLGEHFENDDDVIIAKIDATANELEHTKITSFPTIKLYSKDNQVHDYNGERTLAGLTKFVETDGEGAEPGPSVTEFEEEEDEFVQSFFDGTLKQHLLSEDLPSDWAAKPVKVLVAANFDEVVFDTTKKVLVEFYAPW
Summary
Catalytic Activity
Catalyzes the rearrangement of -S-S- bonds in proteins.
Similarity
Belongs to the protein disulfide isomerase family.
Feature
chain Protein disulfide-isomerase
Uniprot
H9JQN8
Q9GPH2
A0A0K8STM5
A0A437BK29
I4DIQ3
A0A2H1VU96
+ More
A0A0U1VTU3 A0A2A4J7L8 I4DLY7 S4NP33 G9D554 A0A212FP73 A0A1B6G600 U5EXV6 A0A1L8DUB3 A0A1L8DUB0 A0A1B6IVK8 A0A1L8DUD1 V5GZ61 U5EV80 A0A2J7R328 A0A034VPG6 A0A1B6KDW2 A0A146L261 A0A0P5SMK4 A0A0P4WI78 E0VGK5 A0A0N8EAP0 C0JBY4 A0A0P5CNW6 D6WHD6 A0A0P4YEJ8 W8BHI2 A0A1J1I589 A0A1B6DM68 A0A0A9YM47 A0A034VQH7 G9JKY3 A0A0K8TWH4 A0A0P4VT63 A0A0P5JGG6 A0A0P5K4B5 A0A0P5JCL2 E9FUV9 A0A0N8CDX5 A0A0P5QZX1 A0A0P5PBS6 A0A0P5GM21 A0A0P5N6U9 A0A0P5USZ5 A0A067RBI3 A0A0P5VYZ6 A0A0P5JPM2 A0A0P6D8Q6 A0A0P5EXC3 A0A0P5NL58 A0A0P5U7Q0 A0A0P5JJB3 A0A226ECT4 A0A0P5NER3 A0A0P5ENJ2 A0A0P5I7C0 A0A0A1XJL3 A0A2I9LPF6 A0A0N8B9Q8 R4WQK3 A0A0P6HG07 A0A0P5E0R1 A0A0P6DCF2 A0A0P5JJZ6 A0A0P4WUW3 A0A0P5JTS0 A0A3Q0J6F3
A0A0U1VTU3 A0A2A4J7L8 I4DLY7 S4NP33 G9D554 A0A212FP73 A0A1B6G600 U5EXV6 A0A1L8DUB3 A0A1L8DUB0 A0A1B6IVK8 A0A1L8DUD1 V5GZ61 U5EV80 A0A2J7R328 A0A034VPG6 A0A1B6KDW2 A0A146L261 A0A0P5SMK4 A0A0P4WI78 E0VGK5 A0A0N8EAP0 C0JBY4 A0A0P5CNW6 D6WHD6 A0A0P4YEJ8 W8BHI2 A0A1J1I589 A0A1B6DM68 A0A0A9YM47 A0A034VQH7 G9JKY3 A0A0K8TWH4 A0A0P4VT63 A0A0P5JGG6 A0A0P5K4B5 A0A0P5JCL2 E9FUV9 A0A0N8CDX5 A0A0P5QZX1 A0A0P5PBS6 A0A0P5GM21 A0A0P5N6U9 A0A0P5USZ5 A0A067RBI3 A0A0P5VYZ6 A0A0P5JPM2 A0A0P6D8Q6 A0A0P5EXC3 A0A0P5NL58 A0A0P5U7Q0 A0A0P5JJB3 A0A226ECT4 A0A0P5NER3 A0A0P5ENJ2 A0A0P5I7C0 A0A0A1XJL3 A0A2I9LPF6 A0A0N8B9Q8 R4WQK3 A0A0P6HG07 A0A0P5E0R1 A0A0P6DCF2 A0A0P5JJZ6 A0A0P4WUW3 A0A0P5JTS0 A0A3Q0J6F3
EC Number
5.3.4.1
Pubmed
EMBL
BABH01012177
AF325211
AAG45936.1
GBRD01009292
JAG56529.1
RSAL01000043
+ More
RVE50731.1 AK401171 KQ459590 BAM17793.1 KPI97250.1 ODYU01004447 SOQ44326.1 JX183988 AGF69546.1 NWSH01002655 PCG67756.1 AK402305 BAM18927.1 GAIX01012014 JAA80546.1 JF777457 AEU11802.1 AGBW02003612 OWR55527.1 GECZ01011903 JAS57866.1 GANO01000727 JAB59144.1 GFDF01004052 JAV10032.1 GFDF01004053 JAV10031.1 GECU01021031 GECU01016836 GECU01003704 JAS86675.1 JAS90870.1 JAT04003.1 GFDF01004054 JAV10030.1 GALX01002703 JAB65763.1 GANO01001143 JAB58728.1 NEVH01007823 PNF35251.1 GAKP01014588 JAC44364.1 GEBQ01030325 JAT09652.1 GDHC01016151 JAQ02478.1 GDIP01137885 JAL65829.1 GDRN01036013 JAI67374.1 DS235148 EEB12511.1 GDIQ01045348 JAN49389.1 FJ179395 ACN89260.1 GDIP01167789 JAJ55613.1 KQ971321 EFA00647.1 GDIP01228674 JAI94727.1 GAMC01017431 GAMC01017428 JAB89124.1 CVRI01000042 CRK95503.1 GEDC01010527 JAS26771.1 GBHO01009502 GBHO01009500 GBRD01012827 JAG34102.1 JAG34104.1 JAG52999.1 GAKP01014585 JAC44367.1 JN863961 AEW66914.1 GDHF01033678 JAI18636.1 GDKW01002455 JAI54140.1 GDIQ01200294 JAK51431.1 GDIQ01189801 JAK61924.1 GDIQ01200293 JAK51432.1 GL732525 EFX88851.1 GDIP01141247 JAL62467.1 GDIQ01109876 JAL41850.1 GDIQ01130874 JAL20852.1 GDIQ01267948 GDIQ01245936 GDIQ01170947 JAK05789.1 GDIQ01146317 JAL05409.1 GDIP01168587 GDIP01149449 GDIP01112092 GDIP01043389 JAL91622.1 KK852567 KDR21216.1 GDIP01142334 GDIP01094099 JAM09616.1 GDIQ01200295 JAK51430.1 GDIQ01080984 JAN13753.1 GDIQ01265663 JAJ86061.1 GDIQ01143622 JAL08104.1 GDIP01133897 JAL69817.1 GDIQ01198998 JAK52727.1 LNIX01000004 OXA55402.1 GDIQ01143565 JAL08161.1 GDIQ01267549 JAJ84175.1 GDIQ01217681 JAK34044.1 GBXI01002738 JAD11554.1 GFWZ01000275 MBW20265.1 GDIQ01198995 JAK52730.1 AK416967 BAN20182.1 GDIQ01029114 JAN65623.1 GDIP01148149 JAJ75253.1 GDIQ01080983 JAN13754.1 GDIQ01198996 JAK52729.1 GDIP01252819 JAI70582.1 GDIQ01198997 JAK52728.1
RVE50731.1 AK401171 KQ459590 BAM17793.1 KPI97250.1 ODYU01004447 SOQ44326.1 JX183988 AGF69546.1 NWSH01002655 PCG67756.1 AK402305 BAM18927.1 GAIX01012014 JAA80546.1 JF777457 AEU11802.1 AGBW02003612 OWR55527.1 GECZ01011903 JAS57866.1 GANO01000727 JAB59144.1 GFDF01004052 JAV10032.1 GFDF01004053 JAV10031.1 GECU01021031 GECU01016836 GECU01003704 JAS86675.1 JAS90870.1 JAT04003.1 GFDF01004054 JAV10030.1 GALX01002703 JAB65763.1 GANO01001143 JAB58728.1 NEVH01007823 PNF35251.1 GAKP01014588 JAC44364.1 GEBQ01030325 JAT09652.1 GDHC01016151 JAQ02478.1 GDIP01137885 JAL65829.1 GDRN01036013 JAI67374.1 DS235148 EEB12511.1 GDIQ01045348 JAN49389.1 FJ179395 ACN89260.1 GDIP01167789 JAJ55613.1 KQ971321 EFA00647.1 GDIP01228674 JAI94727.1 GAMC01017431 GAMC01017428 JAB89124.1 CVRI01000042 CRK95503.1 GEDC01010527 JAS26771.1 GBHO01009502 GBHO01009500 GBRD01012827 JAG34102.1 JAG34104.1 JAG52999.1 GAKP01014585 JAC44367.1 JN863961 AEW66914.1 GDHF01033678 JAI18636.1 GDKW01002455 JAI54140.1 GDIQ01200294 JAK51431.1 GDIQ01189801 JAK61924.1 GDIQ01200293 JAK51432.1 GL732525 EFX88851.1 GDIP01141247 JAL62467.1 GDIQ01109876 JAL41850.1 GDIQ01130874 JAL20852.1 GDIQ01267948 GDIQ01245936 GDIQ01170947 JAK05789.1 GDIQ01146317 JAL05409.1 GDIP01168587 GDIP01149449 GDIP01112092 GDIP01043389 JAL91622.1 KK852567 KDR21216.1 GDIP01142334 GDIP01094099 JAM09616.1 GDIQ01200295 JAK51430.1 GDIQ01080984 JAN13753.1 GDIQ01265663 JAJ86061.1 GDIQ01143622 JAL08104.1 GDIP01133897 JAL69817.1 GDIQ01198998 JAK52727.1 LNIX01000004 OXA55402.1 GDIQ01143565 JAL08161.1 GDIQ01267549 JAJ84175.1 GDIQ01217681 JAK34044.1 GBXI01002738 JAD11554.1 GFWZ01000275 MBW20265.1 GDIQ01198995 JAK52730.1 AK416967 BAN20182.1 GDIQ01029114 JAN65623.1 GDIP01148149 JAJ75253.1 GDIQ01080983 JAN13754.1 GDIQ01198996 JAK52729.1 GDIP01252819 JAI70582.1 GDIQ01198997 JAK52728.1
Proteomes
Pfam
PF00085 Thioredoxin
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9JQN8
Q9GPH2
A0A0K8STM5
A0A437BK29
I4DIQ3
A0A2H1VU96
+ More
A0A0U1VTU3 A0A2A4J7L8 I4DLY7 S4NP33 G9D554 A0A212FP73 A0A1B6G600 U5EXV6 A0A1L8DUB3 A0A1L8DUB0 A0A1B6IVK8 A0A1L8DUD1 V5GZ61 U5EV80 A0A2J7R328 A0A034VPG6 A0A1B6KDW2 A0A146L261 A0A0P5SMK4 A0A0P4WI78 E0VGK5 A0A0N8EAP0 C0JBY4 A0A0P5CNW6 D6WHD6 A0A0P4YEJ8 W8BHI2 A0A1J1I589 A0A1B6DM68 A0A0A9YM47 A0A034VQH7 G9JKY3 A0A0K8TWH4 A0A0P4VT63 A0A0P5JGG6 A0A0P5K4B5 A0A0P5JCL2 E9FUV9 A0A0N8CDX5 A0A0P5QZX1 A0A0P5PBS6 A0A0P5GM21 A0A0P5N6U9 A0A0P5USZ5 A0A067RBI3 A0A0P5VYZ6 A0A0P5JPM2 A0A0P6D8Q6 A0A0P5EXC3 A0A0P5NL58 A0A0P5U7Q0 A0A0P5JJB3 A0A226ECT4 A0A0P5NER3 A0A0P5ENJ2 A0A0P5I7C0 A0A0A1XJL3 A0A2I9LPF6 A0A0N8B9Q8 R4WQK3 A0A0P6HG07 A0A0P5E0R1 A0A0P6DCF2 A0A0P5JJZ6 A0A0P4WUW3 A0A0P5JTS0 A0A3Q0J6F3
A0A0U1VTU3 A0A2A4J7L8 I4DLY7 S4NP33 G9D554 A0A212FP73 A0A1B6G600 U5EXV6 A0A1L8DUB3 A0A1L8DUB0 A0A1B6IVK8 A0A1L8DUD1 V5GZ61 U5EV80 A0A2J7R328 A0A034VPG6 A0A1B6KDW2 A0A146L261 A0A0P5SMK4 A0A0P4WI78 E0VGK5 A0A0N8EAP0 C0JBY4 A0A0P5CNW6 D6WHD6 A0A0P4YEJ8 W8BHI2 A0A1J1I589 A0A1B6DM68 A0A0A9YM47 A0A034VQH7 G9JKY3 A0A0K8TWH4 A0A0P4VT63 A0A0P5JGG6 A0A0P5K4B5 A0A0P5JCL2 E9FUV9 A0A0N8CDX5 A0A0P5QZX1 A0A0P5PBS6 A0A0P5GM21 A0A0P5N6U9 A0A0P5USZ5 A0A067RBI3 A0A0P5VYZ6 A0A0P5JPM2 A0A0P6D8Q6 A0A0P5EXC3 A0A0P5NL58 A0A0P5U7Q0 A0A0P5JJB3 A0A226ECT4 A0A0P5NER3 A0A0P5ENJ2 A0A0P5I7C0 A0A0A1XJL3 A0A2I9LPF6 A0A0N8B9Q8 R4WQK3 A0A0P6HG07 A0A0P5E0R1 A0A0P6DCF2 A0A0P5JJZ6 A0A0P4WUW3 A0A0P5JTS0 A0A3Q0J6F3
PDB
4EL1
E-value=5.68473e-56,
Score=548
Ontologies
GO
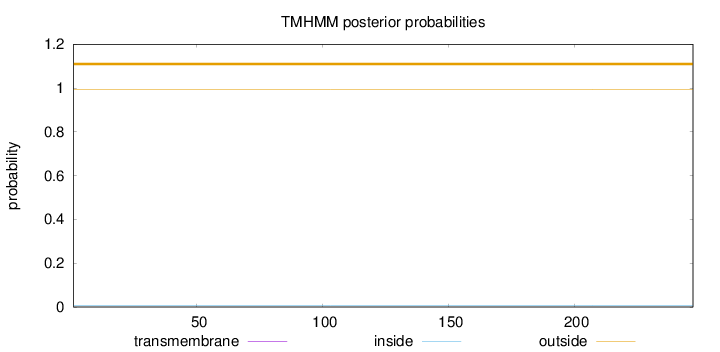
Topology
Length:
247
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00836
outside
1 - 247
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
