Gene
KWMTBOMO16864
Annotation
PREDICTED:_uncharacterized_protein_LOC101743572_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.636 Nuclear Reliability : 1.074
Sequence
CDS
ATGGCACTCCGCGGTGAGTTCCTGAAGGAGACCAAAGTATGCACCATAATATCGCCCCGTCCCGGAACCTCAGCCGACGGAGATGAAGCCCAGGATACGCTTGTCACTAAACAGGGAATTCAAACGGATTTGCTGGACGGAGACGGAACGGTGGACCCTCGGTCCGTGGACCTCTCGCAGCTGCACCAGTCGCGCGTGGGCCTGATCAGCCTCGCGCTGATGCTCATACTGATCGTCTGCTTCCTGGCTTCTGAAACCCTAAGCGAATTCTGGAAGACTAAAAAAAATCCAACTCAAAAATAG
Protein
MALRGEFLKETKVCTIISPRPGTSADGDEAQDTLVTKQGIQTDLLDGDGTVDPRSVDLSQLHQSRVGLISLALMLILIVCFLASETLSEFWKTKKNPTQK
Summary
Uniprot
A0A1E1VY05
A0A194QL88
A0A194QQX4
A0A2A4J7W1
A0A3S2NLL7
A0A0L7LH90
+ More
A0A2J7QQT6 A0A2J7QQS1 E1ZYP8 E2AXV2 A0A026WJH2 A0A067QZ96 A0A151INS8 A0A0J7KUB9 A0A195ECR6 A0A151WPN3 A0A195B0A8 A0A158NXP7 F4WYE3 E2BS63 A0A0P6JLX9 A0A0P5HXX3 A0A0P5WWF8 A0A0N8AZE5 A0A0P5HHG3 A0A0P5EAF1 A0A0P5VE84 A0A0P6CYC6 A0A0P6BQZ2 E0VII0 A0A232EQV4 A0A0N0BHI5 A0A310S771 A0A2A3EHR1 V9IJG0 A0A087ZQA5 A0A423SBC3
A0A2J7QQT6 A0A2J7QQS1 E1ZYP8 E2AXV2 A0A026WJH2 A0A067QZ96 A0A151INS8 A0A0J7KUB9 A0A195ECR6 A0A151WPN3 A0A195B0A8 A0A158NXP7 F4WYE3 E2BS63 A0A0P6JLX9 A0A0P5HXX3 A0A0P5WWF8 A0A0N8AZE5 A0A0P5HHG3 A0A0P5EAF1 A0A0P5VE84 A0A0P6CYC6 A0A0P6BQZ2 E0VII0 A0A232EQV4 A0A0N0BHI5 A0A310S771 A0A2A3EHR1 V9IJG0 A0A087ZQA5 A0A423SBC3
Pubmed
EMBL
GDQN01011454
JAT79600.1
KQ459053
KPJ04206.1
KQ461181
KPJ07749.1
+ More
NWSH01002568 PCG67959.1 RSAL01000042 RVE50782.1 JTDY01001123 KOB74807.1 NEVH01012083 PNF30927.1 PNF30922.1 GL435242 EFN73588.1 GL443736 EFN61716.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 KK853083 KDR11659.1 KQ976914 KYN07095.1 LBMM01003176 KMQ93849.1 KQ979074 KYN23015.1 KQ982851 KYQ49862.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 GL888440 EGI60795.1 GL450151 EFN81455.1 GDIQ01014549 JAN80188.1 GDIQ01226398 JAK25327.1 GDIP01081049 JAM22666.1 GDIQ01227899 JAK23826.1 GDIQ01227898 JAK23827.1 GDIP01144877 JAJ78525.1 GDIP01116333 JAL87381.1 GDIQ01245850 GDIQ01209105 GDIQ01209104 GDIQ01191544 GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GDIP01012624 JAM91091.1 DS235200 EEB13186.1 NNAY01002716 OXU20712.1 KQ435750 KOX76311.1 KQ766333 OAD53704.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 QCYY01004133 ROT61502.1
NWSH01002568 PCG67959.1 RSAL01000042 RVE50782.1 JTDY01001123 KOB74807.1 NEVH01012083 PNF30927.1 PNF30922.1 GL435242 EFN73588.1 GL443736 EFN61716.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 KK853083 KDR11659.1 KQ976914 KYN07095.1 LBMM01003176 KMQ93849.1 KQ979074 KYN23015.1 KQ982851 KYQ49862.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 GL888440 EGI60795.1 GL450151 EFN81455.1 GDIQ01014549 JAN80188.1 GDIQ01226398 JAK25327.1 GDIP01081049 JAM22666.1 GDIQ01227899 JAK23826.1 GDIQ01227898 JAK23827.1 GDIP01144877 JAJ78525.1 GDIP01116333 JAL87381.1 GDIQ01245850 GDIQ01209105 GDIQ01209104 GDIQ01191544 GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GDIP01012624 JAM91091.1 DS235200 EEB13186.1 NNAY01002716 OXU20712.1 KQ435750 KOX76311.1 KQ766333 OAD53704.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 QCYY01004133 ROT61502.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A1E1VY05
A0A194QL88
A0A194QQX4
A0A2A4J7W1
A0A3S2NLL7
A0A0L7LH90
+ More
A0A2J7QQT6 A0A2J7QQS1 E1ZYP8 E2AXV2 A0A026WJH2 A0A067QZ96 A0A151INS8 A0A0J7KUB9 A0A195ECR6 A0A151WPN3 A0A195B0A8 A0A158NXP7 F4WYE3 E2BS63 A0A0P6JLX9 A0A0P5HXX3 A0A0P5WWF8 A0A0N8AZE5 A0A0P5HHG3 A0A0P5EAF1 A0A0P5VE84 A0A0P6CYC6 A0A0P6BQZ2 E0VII0 A0A232EQV4 A0A0N0BHI5 A0A310S771 A0A2A3EHR1 V9IJG0 A0A087ZQA5 A0A423SBC3
A0A2J7QQT6 A0A2J7QQS1 E1ZYP8 E2AXV2 A0A026WJH2 A0A067QZ96 A0A151INS8 A0A0J7KUB9 A0A195ECR6 A0A151WPN3 A0A195B0A8 A0A158NXP7 F4WYE3 E2BS63 A0A0P6JLX9 A0A0P5HXX3 A0A0P5WWF8 A0A0N8AZE5 A0A0P5HHG3 A0A0P5EAF1 A0A0P5VE84 A0A0P6CYC6 A0A0P6BQZ2 E0VII0 A0A232EQV4 A0A0N0BHI5 A0A310S771 A0A2A3EHR1 V9IJG0 A0A087ZQA5 A0A423SBC3
Ontologies
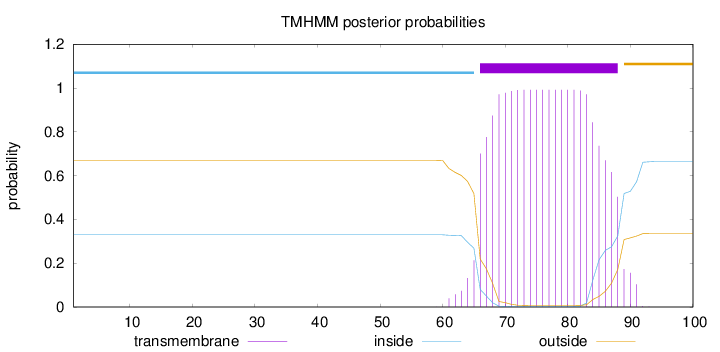
Topology
Length:
100
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.48834
Exp number, first 60 AAs:
0.00144
Total prob of N-in:
0.32975
inside
1 - 65
TMhelix
66 - 88
outside
89 - 100
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
