Gene
KWMTBOMO16824 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004585
Annotation
vitellogenin_precursor_[Bombyx_mori]
Full name
Vitellogenin
Location in the cell
Extracellular Reliability : 1.489 Nuclear Reliability : 1.41
Sequence
CDS
ATGGTGAAACTAGTTTCTGCTGGCATTAACAAAGCTAGAGTGAGAGTTGTGGATTTGAGTGCTAGCTTTGAGGGCAGTCAAGATCAGAATTACGTTTTCACTGGCACCTGGGGTGATAGTCCGGTCGATTCAAAGGTTCAAGGAATGTTATTTGCTGGCACAAACTATCGTTATGCTCATGCGCCTGATCATGTGGAATTCTCCGTCTCATTCCAAGACATGAGTCCTCAATACAGAAACTTCTCCTACCACACATACAGACTCTATGAGTACTTGGGTTACTGGTATACTGAAGCTAATCCTTTGAAACTAACTCAAAATGGTAAAATGGACTTCAAAATTGATTTCTCGTACTTCGATCGTACTTACACCGTCGATATTGCATCCCCATCTGGAGAGGCACGAATGAGGGACATGCCAATTGCAACAATGGCACCTGGTGCTTTGTCTTTCTACCAACCGCTAAAAGCTTATGAACTCGTTGCCAATTACTTCACTGGCCACCAATATCAACCCTACTGTTCTATCGATGGCACAAGGATACATACCTTCAGTAATCGCAGCTACGAATATCCCTATCTCGATCCTGGCACGAGGCAACAGAGAGATCAACAGGAGATCTATATTTCTTACAAGTCTGAGTCAGGACAGGATCTTGAGATCGAAATACAACCAGCGTCTGGAGACTCTGCATATCAAGTGAAAGTCACGACGAATACCAAGAAGATCACCGATGATGACCTCACCATAGGCATCTGTGGACGTATGAGCGGCGAGCAGCGCGACGACTATCTCACTCCAGAAGGTTTGGTCGACAAGCCTGAGCTATATGCCGCAGCCTACAGCCTAAATGAGGAGAACAGTGACCCCAAAACCCAGGAACTGAAGGCTCTAGCAACACAACAAGCTTACTACCCAGAATATAAGTACACAAGCATCCTACGCTCTGACCCAACTTGGCAAGAAGAATCACAGTCCTCGGTGAGGATCAATGGCAGTCTGAAACCGTCTATAAGTCTAGAAGCTACGACAAACACAAGGGCGCTTGTGAAGTTAGGCAACAGGTGCAGTTTTATGAGAACCACGGCGATATCTGTATCACAACTTCGAGAGTCCCCTCGTGTCAGTCGCATTGCCGTGCTGGAGACTACAAAATACAGCACGTTCAAGTGA
Protein
MVKLVSAGINKARVRVVDLSASFEGSQDQNYVFTGTWGDSPVDSKVQGMLFAGTNYRYAHAPDHVEFSVSFQDMSPQYRNFSYHTYRLYEYLGYWYTEANPLKLTQNGKMDFKIDFSYFDRTYTVDIASPSGEARMRDMPIATMAPGALSFYQPLKAYELVANYFTGHQYQPYCSIDGTRIHTFSNRSYEYPYLDPGTRQQRDQQEIYISYKSESGQDLEIEIQPASGDSAYQVKVTTNTKKITDDDLTIGICGRMSGEQRDDYLTPEGLVDKPELYAAAYSLNEENSDPKTQELKALATQQAYYPEYKYTSILRSDPTWQEESQSSVRINGSLKPSISLEATTNTRALVKLGNRCSFMRTTAISVSQLRESPRVSRIAVLETTKYSTFK
Summary
Subunit
Heterotetramer of two heavy and two light chains.
Keywords
Cleavage on pair of basic residues
Complete proteome
Direct protein sequencing
Glycoprotein
Phosphoprotein
Reference proteome
Secreted
Signal
Storage protein
Feature
chain Vitellogenin
Uniprot
H9J4Z5
Q27309
Q9BPP5
Q33CE5
Q9GUX5
Q9BPP7
+ More
A2A0V4 Q59IU2 D3K369 A5A3J2 Q9BPP6 Q59IU3 A0A0N1ICG0 A0A0N1I5M7 A0A0L7LRX8 A0A385HVZ9 A0A0S2C3S6 R4KZ43 A0A2W1BQW6 S4T709 A0A1D5ARX5 A0A0S3G570 W0M288 A0A212ES36 A0A3S2PE51 B0FBK0 A0A059VBJ4 A0A125RTZ3 A0A2H1WVX0 A0A2A4JEQ8 G3LSH9 A7XS62 A0A2A4J2U3 A0A3G1S403 A0A2W1BMM7 A0A2W1BK92 A0A2A4IZG8 A0A2H1W8V4 A0A2A4J1L8 A0A2H1W9B6 A0A2A4JFI3 A8CMX8 A0A2W1BKE7 Q7JPH3 Q25265 Q25269 A0A2H1VRD1 A0A2W1BLH3 A0A2S1XV30 A0A1L2JIS2 A0A2Z6FL27
A2A0V4 Q59IU2 D3K369 A5A3J2 Q9BPP6 Q59IU3 A0A0N1ICG0 A0A0N1I5M7 A0A0L7LRX8 A0A385HVZ9 A0A0S2C3S6 R4KZ43 A0A2W1BQW6 S4T709 A0A1D5ARX5 A0A0S3G570 W0M288 A0A212ES36 A0A3S2PE51 B0FBK0 A0A059VBJ4 A0A125RTZ3 A0A2H1WVX0 A0A2A4JEQ8 G3LSH9 A7XS62 A0A2A4J2U3 A0A3G1S403 A0A2W1BMM7 A0A2W1BK92 A0A2A4IZG8 A0A2H1W8V4 A0A2A4J1L8 A0A2H1W9B6 A0A2A4JFI3 A8CMX8 A0A2W1BKE7 Q7JPH3 Q25265 Q25269 A0A2H1VRD1 A0A2W1BLH3 A0A2S1XV30 A0A1L2JIS2 A0A2Z6FL27
Pubmed
EMBL
BABH01025711
D13160
D30733
AB055845
BAB32642.2
AB239763
+ More
BAE47146.1 EF683091 AB049631 ABS19573.1 BAB16412.1 AB055843 BAB32640.1 AB247378 BAF45319.1 AB190810 BAD91196.1 GU361974 ADB94560.1 EF523567 ABP63663.1 AB055844 BAB32641.1 AB190809 BAD91195.1 KQ459890 KPJ19580.1 KQ458842 KPJ04900.1 JTDY01000218 KOB78233.1 MG799570 AXY55008.1 KT947112 ALN38805.1 JX504706 AGL08685.1 KZ150067 PZC74123.1 JQ723600 AFV40972.1 KT599434 AOH73254.1 KT274176 ALR35190.1 KF501044 AHG29547.1 AGBW02012867 OWR44310.1 RSAL01000078 RVE48729.1 EU290149 ABY56617.1 KJ540279 AHZ89334.1 KT724958 AMD78107.1 ODYU01011461 SOQ57205.1 NWSH01001771 PCG70188.1 JN408698 AEM75020.1 EU095334 ABU68426.1 NWSH01003613 PCG66056.1 MH553377 AXM43802.1 PZC74126.1 PZC74125.1 NWSH01004650 PCG64818.1 ODYU01007081 SOQ49535.1 PCG66057.1 SOQ49536.1 PCG70190.1 EF383143 ABQ23674.1 PZC74124.1 U24161 AAB40975.1 U90756 AAC02818.1 U60186 AAB03336.1 ODYU01003950 SOQ43336.1 PZC74127.1 MF622538 AWJ95280.1 KT892949 AOY34570.1 LC384838 BBE28981.1
BAE47146.1 EF683091 AB049631 ABS19573.1 BAB16412.1 AB055843 BAB32640.1 AB247378 BAF45319.1 AB190810 BAD91196.1 GU361974 ADB94560.1 EF523567 ABP63663.1 AB055844 BAB32641.1 AB190809 BAD91195.1 KQ459890 KPJ19580.1 KQ458842 KPJ04900.1 JTDY01000218 KOB78233.1 MG799570 AXY55008.1 KT947112 ALN38805.1 JX504706 AGL08685.1 KZ150067 PZC74123.1 JQ723600 AFV40972.1 KT599434 AOH73254.1 KT274176 ALR35190.1 KF501044 AHG29547.1 AGBW02012867 OWR44310.1 RSAL01000078 RVE48729.1 EU290149 ABY56617.1 KJ540279 AHZ89334.1 KT724958 AMD78107.1 ODYU01011461 SOQ57205.1 NWSH01001771 PCG70188.1 JN408698 AEM75020.1 EU095334 ABU68426.1 NWSH01003613 PCG66056.1 MH553377 AXM43802.1 PZC74126.1 PZC74125.1 NWSH01004650 PCG64818.1 ODYU01007081 SOQ49535.1 PCG66057.1 SOQ49536.1 PCG70190.1 EF383143 ABQ23674.1 PZC74124.1 U24161 AAB40975.1 U90756 AAC02818.1 U60186 AAB03336.1 ODYU01003950 SOQ43336.1 PZC74127.1 MF622538 AWJ95280.1 KT892949 AOY34570.1 LC384838 BBE28981.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9J4Z5
Q27309
Q9BPP5
Q33CE5
Q9GUX5
Q9BPP7
+ More
A2A0V4 Q59IU2 D3K369 A5A3J2 Q9BPP6 Q59IU3 A0A0N1ICG0 A0A0N1I5M7 A0A0L7LRX8 A0A385HVZ9 A0A0S2C3S6 R4KZ43 A0A2W1BQW6 S4T709 A0A1D5ARX5 A0A0S3G570 W0M288 A0A212ES36 A0A3S2PE51 B0FBK0 A0A059VBJ4 A0A125RTZ3 A0A2H1WVX0 A0A2A4JEQ8 G3LSH9 A7XS62 A0A2A4J2U3 A0A3G1S403 A0A2W1BMM7 A0A2W1BK92 A0A2A4IZG8 A0A2H1W8V4 A0A2A4J1L8 A0A2H1W9B6 A0A2A4JFI3 A8CMX8 A0A2W1BKE7 Q7JPH3 Q25265 Q25269 A0A2H1VRD1 A0A2W1BLH3 A0A2S1XV30 A0A1L2JIS2 A0A2Z6FL27
A2A0V4 Q59IU2 D3K369 A5A3J2 Q9BPP6 Q59IU3 A0A0N1ICG0 A0A0N1I5M7 A0A0L7LRX8 A0A385HVZ9 A0A0S2C3S6 R4KZ43 A0A2W1BQW6 S4T709 A0A1D5ARX5 A0A0S3G570 W0M288 A0A212ES36 A0A3S2PE51 B0FBK0 A0A059VBJ4 A0A125RTZ3 A0A2H1WVX0 A0A2A4JEQ8 G3LSH9 A7XS62 A0A2A4J2U3 A0A3G1S403 A0A2W1BMM7 A0A2W1BK92 A0A2A4IZG8 A0A2H1W8V4 A0A2A4J1L8 A0A2H1W9B6 A0A2A4JFI3 A8CMX8 A0A2W1BKE7 Q7JPH3 Q25265 Q25269 A0A2H1VRD1 A0A2W1BLH3 A0A2S1XV30 A0A1L2JIS2 A0A2Z6FL27
Ontologies
Topology
Subcellular location
Secreted
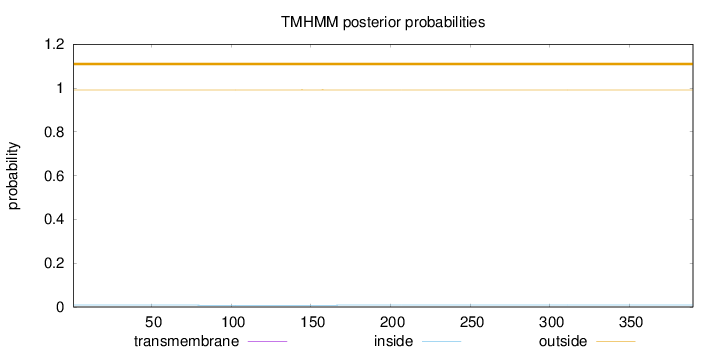
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00269
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00857
outside
1 - 390
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.049826
CSRT
0
Interpretation
Uncertain
