Gene
KWMTBOMO16812 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010204
Annotation
30K_protein_18_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.193 Nuclear Reliability : 1.576
Sequence
CDS
ATGCAGCCAACATACGACTCTACGATATACTGTCTGCCGGTAGTGAGCGAAGGACCACACGAGATGGACCCATTCGACAAGCTCCCCAACTACGTGACCAACGTGGACCTGAAGTATCCGTACTCAGACCTGCCGTACATAGGGCAGTACAAGCTGCTCAAGCTCCCCTTTACTGGAGAGCTGATCGAACATGTCGACTACTGGGGCGAGGGGAGGATCGTCAACGGGGGACTGTATTCGGGCTTCCGCAACTGCTACAACGTAAACCGACAGTACCAAGAGGTCAGCAACGGGCCCGACAAGGGTCGCAAGATCCCCAACAGGATCCCGGTGCGCGACGAGAACGACTGCGACACCCGGGCTTACATCAAGGATGACTCGGTCAAAATAGTGACGCTCATGAGCGCGCCCATCATTCCGAACAAGCGCCAGAGACATTACGAGAATCTAGAGTCCCAGGGTATTAAGCTCCTGGCTGCCGAGCTAAAGAACAAGTTTCTGCTGTACTGTCCCGACTACGAGCTGTCCGCTTACTTACAGGAGCCGACAATGATGGACTCGCACGTGGCTTTCCTCAACAAGCAACTTCTTATGGATCTGCTGTTCAAGTACGTGTCGACCGGGGACTACGACAAGGCGGTGACTATCACGAAGTCCTTGCAAGATGATAACGTCGGCTTTGTTATCAAGGAGCTGGTCGACAGACTCTTACGCGCACGAGAACCTAACCAAGAAAGGGTGAAAATCATCGGAAGATATTACAATCAAGCTCTGAAGCTGGACGCCAATGTCGACAGGTACAACGACCGCCTGGCCTGGGGCGACAGCAAGGACAAGACTAGTCACAGAGTGAGCTGGAAGCTTATCCCGGTATGGGAGAATAACAAGCTGCTGTATAAAATAATGAACACTGAACATACTATGTACCTGAAGCTGGACGTGAACGTGGACGGGTACGGAGACAGGCAAGCGTGGGGCTCCAACAACTCGAACGAGAAGCGCCACCTATGGAAGCTGACGCCTGTAGTGCTGGAGACCGGCAACGTCTTACTCATAGAGAACCACGAGTACGGGCAGAGTCTGAAGCTGGACGCGCACGTGGACAGCTACGGTGACCGCCTGCTCTGGGGAAACAACGGAAACGTCGATGGTAACCCTGGCTACTTCGGCTGGGTCATCAACGCGTGGCCGTAG
Protein
MQPTYDSTIYCLPVVSEGPHEMDPFDKLPNYVTNVDLKYPYSDLPYIGQYKLLKLPFTGELIEHVDYWGEGRIVNGGLYSGFRNCYNVNRQYQEVSNGPDKGRKIPNRIPVRDENDCDTRAYIKDDSVKIVTLMSAPIIPNKRQRHYENLESQGIKLLAAELKNKFLLYCPDYELSAYLQEPTMMDSHVAFLNKQLLMDLLFKYVSTGDYDKAVTITKSLQDDNVGFVIKELVDRLLRAREPNQERVKIIGRYYNQALKLDANVDRYNDRLAWGDSKDKTSHRVSWKLIPVWENNKLLYKIMNTEHTMYLKLDVNVDGYGDRQAWGSNNSNEKRHLWKLTPVVLETGNVLLIENHEYGQSLKLDAHVDSYGDRLLWGNNGNVDGNPGYFGWVINAWP
Summary
Uniprot
EMBL
JN977536
AFC87816.1
BABH01011413
AB436163
BAG70410.1
BABH01026782
+ More
JN977537 AFC87817.1 BABH01036437 BABH01028980 JN977534 AFC87814.1 JN977545 AFC87825.1 JN977540 AFC87820.1 DQ306881 JN977531 ABC18268.1 AFC87811.1 BABH01028979 JN977530 AFC87810.1 JN977541 AFC87821.1 BABH01026781 JN977532 AFC87812.1 KZ149940 PZC76929.1 AB098260 BAC82630.1 NWSH01005147 PCG64400.1 KQ460930 KPJ10718.1 NWSH01000017 PCG80765.1 KZ149893 PZC78918.1 MF497269 AUB45121.1 MG799571 AXY55009.1 NWSH01000685 PCG74802.1 KQ459582 KPI98870.1 PZC78944.1 MF497270 AUB45122.1 ODYU01010790 SOQ56108.1 BABH01022000 PJNJ01000010 PTD20097.1 MAIC01000014 OPB75435.1
JN977537 AFC87817.1 BABH01036437 BABH01028980 JN977534 AFC87814.1 JN977545 AFC87825.1 JN977540 AFC87820.1 DQ306881 JN977531 ABC18268.1 AFC87811.1 BABH01028979 JN977530 AFC87810.1 JN977541 AFC87821.1 BABH01026781 JN977532 AFC87812.1 KZ149940 PZC76929.1 AB098260 BAC82630.1 NWSH01005147 PCG64400.1 KQ460930 KPJ10718.1 NWSH01000017 PCG80765.1 KZ149893 PZC78918.1 MF497269 AUB45121.1 MG799571 AXY55009.1 NWSH01000685 PCG74802.1 KQ459582 KPI98870.1 PZC78944.1 MF497270 AUB45122.1 ODYU01010790 SOQ56108.1 BABH01022000 PJNJ01000010 PTD20097.1 MAIC01000014 OPB75435.1
Proteomes
PRIDE
Pfam
PF03260 Lipoprotein_11
Gene 3D
ProteinModelPortal
PDB
3PUB
E-value=6.42845e-30,
Score=326
Ontologies
GO
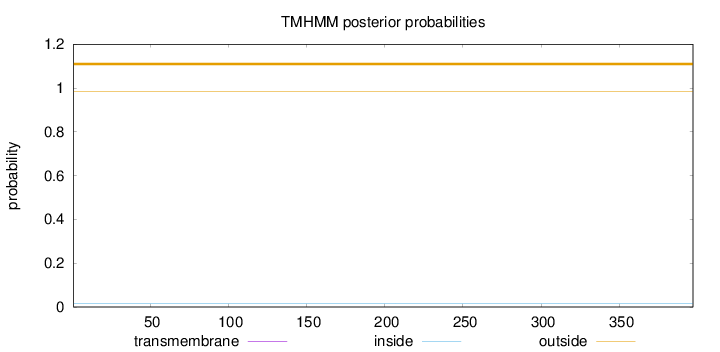
Topology
Length:
397
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000780000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01600
outside
1 - 397
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
