Gene
KWMTBOMO16805
Pre Gene Modal
BGIBMGA008170
Annotation
PREDICTED:_EH_domain-binding_protein_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.852
Sequence
CDS
ATGTCGCAATGGTTCAACCTAGTCAATAAGAAGAACGCACTCCTGAGGAGGCAGATGCAGCTGAACATATTGGAGCAGGAGGAGGACCTGAGCCGCCGCTGCGAGCTGCTGGCGCGCGAGCTGCGCCTCTCCCTCGGCGTCGACGAGTGGAGGAAGACTCCCGGGCAGAAGAGGAGGGAGAGGCTGCTGCTGCAGGAGCTGCTGTCCGCGGTCAACGAGCGAGACCGCCTGGTCCAGGAGATGGACGAGCAGGAGAAAGCGATCGCGGACGACGACGCAATCGAGCGGAACCTGTCGCACGTCGAGATCCAAAGGAAGAACAACTGCATCCTGCAGTAG
Protein
MSQWFNLVNKKNALLRRQMQLNILEQEEDLSRRCELLARELRLSLGVDEWRKTPGQKRRERLLLQELLSAVNERDRLVQEMDEQEKAIADDDAIERNLSHVEIQRKNNCILQ
Summary
Uniprot
A0A437BNG8
A0A2W1B5C6
A0A1E1W293
A0A194Q6Q6
A0A212ENN7
H9JF73
+ More
A0A1B6M5J4 A0A0L7QN88 A0A0T6B2U1 A0A154PF74 A0A0N0BHC9 A0A2A3E2Z2 A0A088A677 A0A310SEI2 A0A026WP25 A0A3L8DWS0 A0A3Q0JE35 A0A3Q0JHU0 A0A1Y1NGF6 A0A1Y1NB72 A0A1Y1NCC5 A0A1W4WJY3 F4WP88 A0A1B6GC02 A0A158NR16 K7IZC5 U4TYW4 A0A195E5N2 A0A151WM95 A0A151I910 U5EXG5 A0A195B135 E2B3V6 A0A1B6LJQ2 A0A1B6L0G6 A0A0A9WE13 A0A195EUB1 E2ADP8 A0A0C9R2M6 A0A1B6JTD0 A0A3Q0J632 A0A336MNP3 A0A336MUK4 A0A336M2T0 A0A336LVB5 A0A232FKL6 A0A2J7R463 A0A2J7R453 A0A2J7R449 A0A2J7R452 A0A224XAX8 A0A0V0G592 A0A069DXH3 N6TUT0 D0IQI2 D6WFM6 A0A1W4UZN9 Q8MR93 A0A1B0DLJ7 B7YZI1 A0A0R1DT98 A0A0J9RFX9 A0A0J9RFX4 A0A1W4UZ96 A1ZAN2 A0A0Q5VLQ9 A4UZL2 A0A1W4UZ88 A0A0J9U468 A0A0Q5VZF0 B3NP34 A0A0R1DTH4 A0A0Q5VVY6 B4P8I8 A0A0Q5VVQ6 B4HMG2 A4UZL3 A0A0R1DT46 A0A0J9REW4 A0A0J9U472 A0A1W4VBZ1 B4QIJ0 A0A1W4VBJ6 A0A0R1DYQ4 A0A1B0GHF6 A0A0A9WJL4 A0A0A9WH21 A0A0P4VWA2 W8C2P7 A0A1L8E5L0 A0A1L8E5S0 A0A1S4G7I6 Q16GZ3 B0WS87 A0A1Q3F311 A0A182GH05 A0A1Q3F2N1 B3MD77 A0A0P8XQD9
A0A1B6M5J4 A0A0L7QN88 A0A0T6B2U1 A0A154PF74 A0A0N0BHC9 A0A2A3E2Z2 A0A088A677 A0A310SEI2 A0A026WP25 A0A3L8DWS0 A0A3Q0JE35 A0A3Q0JHU0 A0A1Y1NGF6 A0A1Y1NB72 A0A1Y1NCC5 A0A1W4WJY3 F4WP88 A0A1B6GC02 A0A158NR16 K7IZC5 U4TYW4 A0A195E5N2 A0A151WM95 A0A151I910 U5EXG5 A0A195B135 E2B3V6 A0A1B6LJQ2 A0A1B6L0G6 A0A0A9WE13 A0A195EUB1 E2ADP8 A0A0C9R2M6 A0A1B6JTD0 A0A3Q0J632 A0A336MNP3 A0A336MUK4 A0A336M2T0 A0A336LVB5 A0A232FKL6 A0A2J7R463 A0A2J7R453 A0A2J7R449 A0A2J7R452 A0A224XAX8 A0A0V0G592 A0A069DXH3 N6TUT0 D0IQI2 D6WFM6 A0A1W4UZN9 Q8MR93 A0A1B0DLJ7 B7YZI1 A0A0R1DT98 A0A0J9RFX9 A0A0J9RFX4 A0A1W4UZ96 A1ZAN2 A0A0Q5VLQ9 A4UZL2 A0A1W4UZ88 A0A0J9U468 A0A0Q5VZF0 B3NP34 A0A0R1DTH4 A0A0Q5VVY6 B4P8I8 A0A0Q5VVQ6 B4HMG2 A4UZL3 A0A0R1DT46 A0A0J9REW4 A0A0J9U472 A0A1W4VBZ1 B4QIJ0 A0A1W4VBJ6 A0A0R1DYQ4 A0A1B0GHF6 A0A0A9WJL4 A0A0A9WH21 A0A0P4VWA2 W8C2P7 A0A1L8E5L0 A0A1L8E5S0 A0A1S4G7I6 Q16GZ3 B0WS87 A0A1Q3F311 A0A182GH05 A0A1Q3F2N1 B3MD77 A0A0P8XQD9
Pubmed
28756777
26354079
22118469
19121390
24508170
30249741
+ More
28004739 21719571 21347285 20075255 23537049 20798317 25401762 28648823 26334808 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 22936249 26109357 26109356 18057021 27129103 24495485 17510324 26483478
28004739 21719571 21347285 20075255 23537049 20798317 25401762 28648823 26334808 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 22936249 26109357 26109356 18057021 27129103 24495485 17510324 26483478
EMBL
RSAL01000028
RVE51898.1
KZ150393
PZC71038.1
GDQN01011159
GDQN01010000
+ More
GDQN01000303 JAT79895.1 JAT81054.1 JAT90751.1 KQ459386 KPJ01227.1 AGBW02013627 OWR43093.1 BABH01026837 BABH01026838 GEBQ01008782 JAT31195.1 KQ414856 KOC60095.1 LJIG01016247 KRT81175.1 KQ434879 KZC09958.1 KQ435757 KOX75872.1 KZ288430 PBC25844.1 KQ760662 OAD59537.1 KK107139 EZA57698.1 QOIP01000003 RLU24887.1 GEZM01007454 JAV95186.1 GEZM01007456 JAV95184.1 GEZM01007458 GEZM01007457 GEZM01007455 GEZM01007453 GEZM01007452 JAV95188.1 GL888243 EGI64069.1 GECZ01009802 JAS59967.1 ADTU01023744 KB631866 ERL86809.1 KQ979608 KYN20473.1 KQ982944 KYQ49012.1 KQ978324 KYM95107.1 GANO01002547 JAB57324.1 KQ976691 KYM77914.1 GL445407 EFN89625.1 GEBQ01016062 JAT23915.1 GEBQ01022832 JAT17145.1 GBHO01036912 JAG06692.1 KQ981965 KYN31840.1 GL438820 EFN68410.1 GBYB01002219 GBYB01002220 JAG71986.1 JAG71987.1 GECU01005307 JAT02400.1 UFQS01001832 UFQT01001832 SSX12483.1 SSX31926.1 SSX12484.1 SSX31927.1 UFQS01000115 UFQT01000115 SSW99898.1 SSX20278.1 SSW99897.1 SSX20277.1 NNAY01000112 OXU30867.1 NEVH01007815 PNF35624.1 PNF35622.1 PNF35621.1 PNF35623.1 GFTR01008332 JAW08094.1 GECL01002907 JAP03217.1 GBGD01000363 JAC88526.1 APGK01017648 KB740047 ENN81833.1 BT100220 ACY46095.1 KQ971319 EFA00250.2 AY121714 AAM52041.1 AJVK01036445 AJVK01036446 AJVK01036447 AE013599 ACL83137.2 CM000158 KRK00392.1 CM002911 KMY94434.1 KMY94437.1 KMY94429.1 BT099671 AAF57918.3 ACV32771.1 CH954179 KQS62390.1 KQS62391.1 AAS64823.1 KMY94430.1 KQS62394.1 EDV55673.1 KQS62396.1 KRK00389.1 KQS62392.1 KQS62395.1 EDW92205.1 KRK00393.1 KQS62393.1 CH480816 EDW48230.1 AAS64824.2 KRK00390.1 KRK00391.1 KMY94436.1 KMY94435.1 CM000362 EDX07437.1 KMY94428.1 KRK00394.1 AJWK01004552 GBHO01036911 GBRD01001155 JAG06693.1 JAG64666.1 GBHO01036913 GBRD01001153 JAG06691.1 JAG64668.1 GDKW01000208 JAI56387.1 GAMC01010536 JAB96019.1 GFDF01000091 JAV13993.1 GFDF01000090 JAV13994.1 CH478221 EAT33515.1 DS232066 EDS33714.1 GFDL01013105 JAV21940.1 JXUM01062708 JXUM01062709 JXUM01062710 GFDL01013229 JAV21816.1 CH902619 EDV37410.1 KPU76803.1
GDQN01000303 JAT79895.1 JAT81054.1 JAT90751.1 KQ459386 KPJ01227.1 AGBW02013627 OWR43093.1 BABH01026837 BABH01026838 GEBQ01008782 JAT31195.1 KQ414856 KOC60095.1 LJIG01016247 KRT81175.1 KQ434879 KZC09958.1 KQ435757 KOX75872.1 KZ288430 PBC25844.1 KQ760662 OAD59537.1 KK107139 EZA57698.1 QOIP01000003 RLU24887.1 GEZM01007454 JAV95186.1 GEZM01007456 JAV95184.1 GEZM01007458 GEZM01007457 GEZM01007455 GEZM01007453 GEZM01007452 JAV95188.1 GL888243 EGI64069.1 GECZ01009802 JAS59967.1 ADTU01023744 KB631866 ERL86809.1 KQ979608 KYN20473.1 KQ982944 KYQ49012.1 KQ978324 KYM95107.1 GANO01002547 JAB57324.1 KQ976691 KYM77914.1 GL445407 EFN89625.1 GEBQ01016062 JAT23915.1 GEBQ01022832 JAT17145.1 GBHO01036912 JAG06692.1 KQ981965 KYN31840.1 GL438820 EFN68410.1 GBYB01002219 GBYB01002220 JAG71986.1 JAG71987.1 GECU01005307 JAT02400.1 UFQS01001832 UFQT01001832 SSX12483.1 SSX31926.1 SSX12484.1 SSX31927.1 UFQS01000115 UFQT01000115 SSW99898.1 SSX20278.1 SSW99897.1 SSX20277.1 NNAY01000112 OXU30867.1 NEVH01007815 PNF35624.1 PNF35622.1 PNF35621.1 PNF35623.1 GFTR01008332 JAW08094.1 GECL01002907 JAP03217.1 GBGD01000363 JAC88526.1 APGK01017648 KB740047 ENN81833.1 BT100220 ACY46095.1 KQ971319 EFA00250.2 AY121714 AAM52041.1 AJVK01036445 AJVK01036446 AJVK01036447 AE013599 ACL83137.2 CM000158 KRK00392.1 CM002911 KMY94434.1 KMY94437.1 KMY94429.1 BT099671 AAF57918.3 ACV32771.1 CH954179 KQS62390.1 KQS62391.1 AAS64823.1 KMY94430.1 KQS62394.1 EDV55673.1 KQS62396.1 KRK00389.1 KQS62392.1 KQS62395.1 EDW92205.1 KRK00393.1 KQS62393.1 CH480816 EDW48230.1 AAS64824.2 KRK00390.1 KRK00391.1 KMY94436.1 KMY94435.1 CM000362 EDX07437.1 KMY94428.1 KRK00394.1 AJWK01004552 GBHO01036911 GBRD01001155 JAG06693.1 JAG64666.1 GBHO01036913 GBRD01001153 JAG06691.1 JAG64668.1 GDKW01000208 JAI56387.1 GAMC01010536 JAB96019.1 GFDF01000091 JAV13993.1 GFDF01000090 JAV13994.1 CH478221 EAT33515.1 DS232066 EDS33714.1 GFDL01013105 JAV21940.1 JXUM01062708 JXUM01062709 JXUM01062710 GFDL01013229 JAV21816.1 CH902619 EDV37410.1 KPU76803.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000005204
UP000053825
UP000076502
+ More
UP000053105 UP000242457 UP000005203 UP000053097 UP000279307 UP000079169 UP000192223 UP000007755 UP000005205 UP000002358 UP000030742 UP000078492 UP000075809 UP000078542 UP000078540 UP000008237 UP000078541 UP000000311 UP000215335 UP000235965 UP000019118 UP000007266 UP000192221 UP000092462 UP000000803 UP000002282 UP000008711 UP000001292 UP000000304 UP000092461 UP000008820 UP000002320 UP000069940 UP000007801
UP000053105 UP000242457 UP000005203 UP000053097 UP000279307 UP000079169 UP000192223 UP000007755 UP000005205 UP000002358 UP000030742 UP000078492 UP000075809 UP000078542 UP000078540 UP000008237 UP000078541 UP000000311 UP000215335 UP000235965 UP000019118 UP000007266 UP000192221 UP000092462 UP000000803 UP000002282 UP000008711 UP000001292 UP000000304 UP000092461 UP000008820 UP000002320 UP000069940 UP000007801
Interpro
SUPFAM
SSF47576
SSF47576
Gene 3D
CDD
ProteinModelPortal
A0A437BNG8
A0A2W1B5C6
A0A1E1W293
A0A194Q6Q6
A0A212ENN7
H9JF73
+ More
A0A1B6M5J4 A0A0L7QN88 A0A0T6B2U1 A0A154PF74 A0A0N0BHC9 A0A2A3E2Z2 A0A088A677 A0A310SEI2 A0A026WP25 A0A3L8DWS0 A0A3Q0JE35 A0A3Q0JHU0 A0A1Y1NGF6 A0A1Y1NB72 A0A1Y1NCC5 A0A1W4WJY3 F4WP88 A0A1B6GC02 A0A158NR16 K7IZC5 U4TYW4 A0A195E5N2 A0A151WM95 A0A151I910 U5EXG5 A0A195B135 E2B3V6 A0A1B6LJQ2 A0A1B6L0G6 A0A0A9WE13 A0A195EUB1 E2ADP8 A0A0C9R2M6 A0A1B6JTD0 A0A3Q0J632 A0A336MNP3 A0A336MUK4 A0A336M2T0 A0A336LVB5 A0A232FKL6 A0A2J7R463 A0A2J7R453 A0A2J7R449 A0A2J7R452 A0A224XAX8 A0A0V0G592 A0A069DXH3 N6TUT0 D0IQI2 D6WFM6 A0A1W4UZN9 Q8MR93 A0A1B0DLJ7 B7YZI1 A0A0R1DT98 A0A0J9RFX9 A0A0J9RFX4 A0A1W4UZ96 A1ZAN2 A0A0Q5VLQ9 A4UZL2 A0A1W4UZ88 A0A0J9U468 A0A0Q5VZF0 B3NP34 A0A0R1DTH4 A0A0Q5VVY6 B4P8I8 A0A0Q5VVQ6 B4HMG2 A4UZL3 A0A0R1DT46 A0A0J9REW4 A0A0J9U472 A0A1W4VBZ1 B4QIJ0 A0A1W4VBJ6 A0A0R1DYQ4 A0A1B0GHF6 A0A0A9WJL4 A0A0A9WH21 A0A0P4VWA2 W8C2P7 A0A1L8E5L0 A0A1L8E5S0 A0A1S4G7I6 Q16GZ3 B0WS87 A0A1Q3F311 A0A182GH05 A0A1Q3F2N1 B3MD77 A0A0P8XQD9
A0A1B6M5J4 A0A0L7QN88 A0A0T6B2U1 A0A154PF74 A0A0N0BHC9 A0A2A3E2Z2 A0A088A677 A0A310SEI2 A0A026WP25 A0A3L8DWS0 A0A3Q0JE35 A0A3Q0JHU0 A0A1Y1NGF6 A0A1Y1NB72 A0A1Y1NCC5 A0A1W4WJY3 F4WP88 A0A1B6GC02 A0A158NR16 K7IZC5 U4TYW4 A0A195E5N2 A0A151WM95 A0A151I910 U5EXG5 A0A195B135 E2B3V6 A0A1B6LJQ2 A0A1B6L0G6 A0A0A9WE13 A0A195EUB1 E2ADP8 A0A0C9R2M6 A0A1B6JTD0 A0A3Q0J632 A0A336MNP3 A0A336MUK4 A0A336M2T0 A0A336LVB5 A0A232FKL6 A0A2J7R463 A0A2J7R453 A0A2J7R449 A0A2J7R452 A0A224XAX8 A0A0V0G592 A0A069DXH3 N6TUT0 D0IQI2 D6WFM6 A0A1W4UZN9 Q8MR93 A0A1B0DLJ7 B7YZI1 A0A0R1DT98 A0A0J9RFX9 A0A0J9RFX4 A0A1W4UZ96 A1ZAN2 A0A0Q5VLQ9 A4UZL2 A0A1W4UZ88 A0A0J9U468 A0A0Q5VZF0 B3NP34 A0A0R1DTH4 A0A0Q5VVY6 B4P8I8 A0A0Q5VVQ6 B4HMG2 A4UZL3 A0A0R1DT46 A0A0J9REW4 A0A0J9U472 A0A1W4VBZ1 B4QIJ0 A0A1W4VBJ6 A0A0R1DYQ4 A0A1B0GHF6 A0A0A9WJL4 A0A0A9WH21 A0A0P4VWA2 W8C2P7 A0A1L8E5L0 A0A1L8E5S0 A0A1S4G7I6 Q16GZ3 B0WS87 A0A1Q3F311 A0A182GH05 A0A1Q3F2N1 B3MD77 A0A0P8XQD9
Ontologies
GO
PANTHER
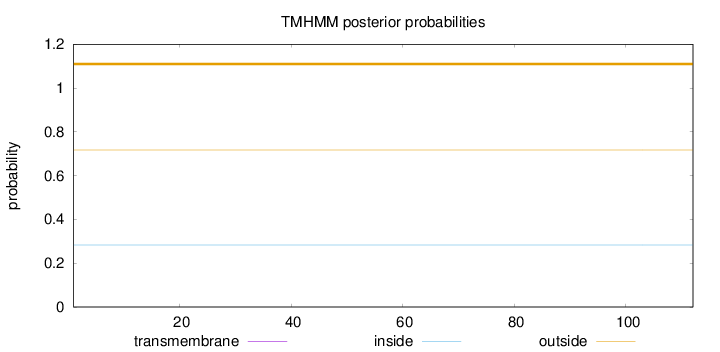
Topology
Length:
112
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.28299
outside
1 - 112
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
