Gene
KWMTBOMO16797
Annotation
uncharacterized_protein_Dyak_GE27431_[Drosophila_yakuba]
Location in the cell
Cytoplasmic Reliability : 1.547 Nuclear Reliability : 2.063
Sequence
CDS
ATGAAAGGAGCGCAGATCTCGACTCCCATGGAACTGGTGCAGGCTACGACGGCAGCGTGTGACGCTTCTATGCCAAAGCAAAACAACCGCAAGAAGAAAGGAGTCTACTGGTGGAACAAGGAAATTGCGGAAGCAAGGAAGGAGTGTTTGAAATGCCGAAGGAGGGAAAGTAACGAAGAAAATGAACACAAAAAGCAGTATAAGGAAGCGAAACTGGAGCTACAAAGACGAATATGGAAAGCGAAAGACGATAAATGGAGGGAAGTGTGCGAGGAAGTCGACAATGATATATGGGGACTGGGATACAAAAATAGTGTCAAAAAACTACGCGGAAACTCACCGATAAATATGGGCATGGCCCAAATTCAAAGCATAGCTGATGGCCTCTTCCCGGAACACATAGATTTCGAACGGAAAACGAAACCATCACAGGAGTTCCCAGAGCCCAGGGCCCGACCGATTCCGCCGGAGATAGTGACTCTAGCAGTAGCAGTAATTCCAGACAGGATAAGGCTTGTGTTGCAGAAGGTGCTGGAGTCTGGCGTCTTCCCCAACAGCTGGAAAACAGCCAGACTAGTCCTGCTAAGAAAACCAGGCAAAACCGCTGCTGAAGATCCTGCGGGGCACAGGCCGCTATGTCTACTGGACTGCTACGGAAAGCTGCTGGAGTCGCTGATCCTAGCAGAATAG
Protein
MKGAQISTPMELVQATTAACDASMPKQNNRKKKGVYWWNKEIAEARKECLKCRRRESNEENEHKKQYKEAKLELQRRIWKAKDDKWREVCEEVDNDIWGLGYKNSVKKLRGNSPINMGMAQIQSIADGLFPEHIDFERKTKPSQEFPEPRARPIPPEIVTLAVAVIPDRIRLVLQKVLESGVFPNSWKTARLVLLRKPGKTAAEDPAGHRPLCLLDCYGKLLESLILAE
Summary
Uniprot
J9KUM6
J9M402
A0A0R1E8T3
A0A139W9A6
F7IYV2
A0A0K8S5I5
+ More
J9KTA9 A0A139WN93 F7IYV7 A0A139WNC9 A0A139W8G2 J9KNS1 A0A139WA31 J9K6F0 A0A139WAH4 A0A139W9B3 A0A2M4BB78 A0A2M4BB89 A0A2S2P488 J9L6Z9 W8ANK7 W8ADE4 V5GYB3 F7IYV5 A0A1Y1KQU0 A0A2S2NJ92 A0A2M4BBA0 A0A2M3Z1V8 A0A2M3Z1C1 A0A2M4AK35 A0A142LX43 W8AMZ9 A0A2M4CKQ7 J9LVU7 J9KE62 A0A1W7R6I9 X1WNG1 A0A2M4BC28 A0A023EW96 A0A0A9Z010 A0A2M4BDI3 J9KT70 A0A2M4CS98 A0A2M4CL98 A0A1W7R6C5 A0A224XIZ2 A0A2M4BCE1 A0A1S4EMG7 T1DCM0 A0A2M4AGH2 A0A2M4AHG6 J9LJ80 A0A0A9ZCY0 J9KNN8 A0A2M4ADW9 K7JP42 A0A2M4BC24 J9LA43 A0A2M4BC18 A0A2M4BBV9 A0A2M4BC01 A0A2H8TR36 A0A139W9J0 T1DG89 A0A0J7KQY4 A0A2S2N8I2 T1DG59 D6WC19 A0A0K8VPH1 J9KJG8 A0A0J7NJI3 X1X2B5 A0A2S2NQQ6 V9GZD3 J9JWJ3 J9LPN2 A0A139W907 J9JY94 A0A142LX45 K7JSS0 A0A2S2PEZ7 J9M5P2 J9L0J0 A0A034WR48 Q868S4 X1WJY4 A0A0K8TR60 A0A2S2PKY3 A0A2S2NGH7 A0A0J7K1Q7 J9LM37 X1WZ64 J9KM84 F7IYU8 A0A139W940 X1WWY8 X1XT97 A0A0J7NAV8 A0A0J7KIY5 A0A2S2NJ82
J9KTA9 A0A139WN93 F7IYV7 A0A139WNC9 A0A139W8G2 J9KNS1 A0A139WA31 J9K6F0 A0A139WAH4 A0A139W9B3 A0A2M4BB78 A0A2M4BB89 A0A2S2P488 J9L6Z9 W8ANK7 W8ADE4 V5GYB3 F7IYV5 A0A1Y1KQU0 A0A2S2NJ92 A0A2M4BBA0 A0A2M3Z1V8 A0A2M3Z1C1 A0A2M4AK35 A0A142LX43 W8AMZ9 A0A2M4CKQ7 J9LVU7 J9KE62 A0A1W7R6I9 X1WNG1 A0A2M4BC28 A0A023EW96 A0A0A9Z010 A0A2M4BDI3 J9KT70 A0A2M4CS98 A0A2M4CL98 A0A1W7R6C5 A0A224XIZ2 A0A2M4BCE1 A0A1S4EMG7 T1DCM0 A0A2M4AGH2 A0A2M4AHG6 J9LJ80 A0A0A9ZCY0 J9KNN8 A0A2M4ADW9 K7JP42 A0A2M4BC24 J9LA43 A0A2M4BC18 A0A2M4BBV9 A0A2M4BC01 A0A2H8TR36 A0A139W9J0 T1DG89 A0A0J7KQY4 A0A2S2N8I2 T1DG59 D6WC19 A0A0K8VPH1 J9KJG8 A0A0J7NJI3 X1X2B5 A0A2S2NQQ6 V9GZD3 J9JWJ3 J9LPN2 A0A139W907 J9JY94 A0A142LX45 K7JSS0 A0A2S2PEZ7 J9M5P2 J9L0J0 A0A034WR48 Q868S4 X1WJY4 A0A0K8TR60 A0A2S2PKY3 A0A2S2NGH7 A0A0J7K1Q7 J9LM37 X1WZ64 J9KM84 F7IYU8 A0A139W940 X1WWY8 X1XT97 A0A0J7NAV8 A0A0J7KIY5 A0A2S2NJ82
Pubmed
EMBL
ABLF02041557
ABLF02028779
CH892793
KRK05561.1
KQ972544
KXZ75870.1
+ More
AB593323 BAK38643.1 GBRD01017378 JAG48449.1 ABLF02009502 KQ971310 KYB29499.1 AB593327 BAK38648.1 KYB29500.1 KQ973343 KXZ75566.1 ABLF02018808 KQ971675 KYB24771.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 KQ971388 KYB24913.1 KXZ75869.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 GGMR01011682 MBY24301.1 ABLF02021100 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 GALX01003058 JAB65408.1 AB593325 BAK38646.1 GEZM01076296 JAV63782.1 GGMR01004227 MBY16846.1 GGFJ01001166 MBW50307.1 GGFM01001733 MBW22484.1 GGFM01001554 MBW22305.1 GGFK01007771 MBW41092.1 KU543682 AMS38369.1 GAMC01019173 JAB87382.1 GGFL01001583 MBW65761.1 ABLF02034925 ABLF02007249 GEHC01000915 JAV46730.1 ABLF02019087 ABLF02019088 ABLF02026448 ABLF02049713 GGFJ01001439 MBW50580.1 GAPW01000192 JAC13406.1 GBHO01006398 JAG37206.1 GGFJ01001727 MBW50868.1 ABLF02010223 GGFL01004044 MBW68222.1 GGFL01001916 MBW66094.1 GEHC01000964 JAV46681.1 GFTR01008427 JAW07999.1 GGFJ01001541 MBW50682.1 GALA01001777 JAA93075.1 GGFK01006563 MBW39884.1 GGFK01006866 MBW40187.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GBHO01002406 JAG41198.1 ABLF02023257 GGFK01005660 MBW38981.1 AAZX01012143 AAZX01012194 GGFJ01001454 MBW50595.1 ABLF02018098 GGFJ01001453 MBW50594.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 GFXV01003903 MBW15708.1 KQ972226 KYB24577.1 GALA01001776 JAA93076.1 LBMM01004262 KMQ92649.1 GGMR01000779 MBY13398.1 GALA01000302 JAA94550.1 KQ971309 EEZ99146.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 ABLF02041886 LBMM01004261 KMQ92650.1 ABLF02019402 ABLF02059310 GGMR01006838 MBY19457.1 M93690 AAA29363.1 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02038060 ABLF02038061 ABLF02058893 KQ972774 KXZ75780.1 ABLF02007989 KU543683 AMS38371.1 AAZX01003982 AAZX01005071 AAZX01005686 AAZX01011269 AAZX01017893 AAZX01026102 GGMR01015159 MBY27778.1 ABLF02010355 ABLF02027248 GAKP01002292 JAC56660.1 AB090815 BAC57906.1 ABLF02011238 GDAI01000741 JAI16862.1 GGMR01017503 MBY30122.1 GGMR01003651 MBY16270.1 LBMM01017390 KMQ84101.1 ABLF02014650 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 AB593321 BAK38639.1 KQ972691 KXZ75806.1 ABLF02027243 ABLF02035831 ABLF02050978 ABLF02062611 ABLF02041320 LBMM01007425 KMQ89740.1 LBMM01006954 KMQ90166.1 GGMR01004600 MBY17219.1
AB593323 BAK38643.1 GBRD01017378 JAG48449.1 ABLF02009502 KQ971310 KYB29499.1 AB593327 BAK38648.1 KYB29500.1 KQ973343 KXZ75566.1 ABLF02018808 KQ971675 KYB24771.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 KQ971388 KYB24913.1 KXZ75869.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 GGMR01011682 MBY24301.1 ABLF02021100 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 GALX01003058 JAB65408.1 AB593325 BAK38646.1 GEZM01076296 JAV63782.1 GGMR01004227 MBY16846.1 GGFJ01001166 MBW50307.1 GGFM01001733 MBW22484.1 GGFM01001554 MBW22305.1 GGFK01007771 MBW41092.1 KU543682 AMS38369.1 GAMC01019173 JAB87382.1 GGFL01001583 MBW65761.1 ABLF02034925 ABLF02007249 GEHC01000915 JAV46730.1 ABLF02019087 ABLF02019088 ABLF02026448 ABLF02049713 GGFJ01001439 MBW50580.1 GAPW01000192 JAC13406.1 GBHO01006398 JAG37206.1 GGFJ01001727 MBW50868.1 ABLF02010223 GGFL01004044 MBW68222.1 GGFL01001916 MBW66094.1 GEHC01000964 JAV46681.1 GFTR01008427 JAW07999.1 GGFJ01001541 MBW50682.1 GALA01001777 JAA93075.1 GGFK01006563 MBW39884.1 GGFK01006866 MBW40187.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GBHO01002406 JAG41198.1 ABLF02023257 GGFK01005660 MBW38981.1 AAZX01012143 AAZX01012194 GGFJ01001454 MBW50595.1 ABLF02018098 GGFJ01001453 MBW50594.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 GFXV01003903 MBW15708.1 KQ972226 KYB24577.1 GALA01001776 JAA93076.1 LBMM01004262 KMQ92649.1 GGMR01000779 MBY13398.1 GALA01000302 JAA94550.1 KQ971309 EEZ99146.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 ABLF02041886 LBMM01004261 KMQ92650.1 ABLF02019402 ABLF02059310 GGMR01006838 MBY19457.1 M93690 AAA29363.1 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02038060 ABLF02038061 ABLF02058893 KQ972774 KXZ75780.1 ABLF02007989 KU543683 AMS38371.1 AAZX01003982 AAZX01005071 AAZX01005686 AAZX01011269 AAZX01017893 AAZX01026102 GGMR01015159 MBY27778.1 ABLF02010355 ABLF02027248 GAKP01002292 JAC56660.1 AB090815 BAC57906.1 ABLF02011238 GDAI01000741 JAI16862.1 GGMR01017503 MBY30122.1 GGMR01003651 MBY16270.1 LBMM01017390 KMQ84101.1 ABLF02014650 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 AB593321 BAK38639.1 KQ972691 KXZ75806.1 ABLF02027243 ABLF02035831 ABLF02050978 ABLF02062611 ABLF02041320 LBMM01007425 KMQ89740.1 LBMM01006954 KMQ90166.1 GGMR01004600 MBY17219.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
J9KUM6
J9M402
A0A0R1E8T3
A0A139W9A6
F7IYV2
A0A0K8S5I5
+ More
J9KTA9 A0A139WN93 F7IYV7 A0A139WNC9 A0A139W8G2 J9KNS1 A0A139WA31 J9K6F0 A0A139WAH4 A0A139W9B3 A0A2M4BB78 A0A2M4BB89 A0A2S2P488 J9L6Z9 W8ANK7 W8ADE4 V5GYB3 F7IYV5 A0A1Y1KQU0 A0A2S2NJ92 A0A2M4BBA0 A0A2M3Z1V8 A0A2M3Z1C1 A0A2M4AK35 A0A142LX43 W8AMZ9 A0A2M4CKQ7 J9LVU7 J9KE62 A0A1W7R6I9 X1WNG1 A0A2M4BC28 A0A023EW96 A0A0A9Z010 A0A2M4BDI3 J9KT70 A0A2M4CS98 A0A2M4CL98 A0A1W7R6C5 A0A224XIZ2 A0A2M4BCE1 A0A1S4EMG7 T1DCM0 A0A2M4AGH2 A0A2M4AHG6 J9LJ80 A0A0A9ZCY0 J9KNN8 A0A2M4ADW9 K7JP42 A0A2M4BC24 J9LA43 A0A2M4BC18 A0A2M4BBV9 A0A2M4BC01 A0A2H8TR36 A0A139W9J0 T1DG89 A0A0J7KQY4 A0A2S2N8I2 T1DG59 D6WC19 A0A0K8VPH1 J9KJG8 A0A0J7NJI3 X1X2B5 A0A2S2NQQ6 V9GZD3 J9JWJ3 J9LPN2 A0A139W907 J9JY94 A0A142LX45 K7JSS0 A0A2S2PEZ7 J9M5P2 J9L0J0 A0A034WR48 Q868S4 X1WJY4 A0A0K8TR60 A0A2S2PKY3 A0A2S2NGH7 A0A0J7K1Q7 J9LM37 X1WZ64 J9KM84 F7IYU8 A0A139W940 X1WWY8 X1XT97 A0A0J7NAV8 A0A0J7KIY5 A0A2S2NJ82
J9KTA9 A0A139WN93 F7IYV7 A0A139WNC9 A0A139W8G2 J9KNS1 A0A139WA31 J9K6F0 A0A139WAH4 A0A139W9B3 A0A2M4BB78 A0A2M4BB89 A0A2S2P488 J9L6Z9 W8ANK7 W8ADE4 V5GYB3 F7IYV5 A0A1Y1KQU0 A0A2S2NJ92 A0A2M4BBA0 A0A2M3Z1V8 A0A2M3Z1C1 A0A2M4AK35 A0A142LX43 W8AMZ9 A0A2M4CKQ7 J9LVU7 J9KE62 A0A1W7R6I9 X1WNG1 A0A2M4BC28 A0A023EW96 A0A0A9Z010 A0A2M4BDI3 J9KT70 A0A2M4CS98 A0A2M4CL98 A0A1W7R6C5 A0A224XIZ2 A0A2M4BCE1 A0A1S4EMG7 T1DCM0 A0A2M4AGH2 A0A2M4AHG6 J9LJ80 A0A0A9ZCY0 J9KNN8 A0A2M4ADW9 K7JP42 A0A2M4BC24 J9LA43 A0A2M4BC18 A0A2M4BBV9 A0A2M4BC01 A0A2H8TR36 A0A139W9J0 T1DG89 A0A0J7KQY4 A0A2S2N8I2 T1DG59 D6WC19 A0A0K8VPH1 J9KJG8 A0A0J7NJI3 X1X2B5 A0A2S2NQQ6 V9GZD3 J9JWJ3 J9LPN2 A0A139W907 J9JY94 A0A142LX45 K7JSS0 A0A2S2PEZ7 J9M5P2 J9L0J0 A0A034WR48 Q868S4 X1WJY4 A0A0K8TR60 A0A2S2PKY3 A0A2S2NGH7 A0A0J7K1Q7 J9LM37 X1WZ64 J9KM84 F7IYU8 A0A139W940 X1WWY8 X1XT97 A0A0J7NAV8 A0A0J7KIY5 A0A2S2NJ82
Ontologies
KEGG
GO
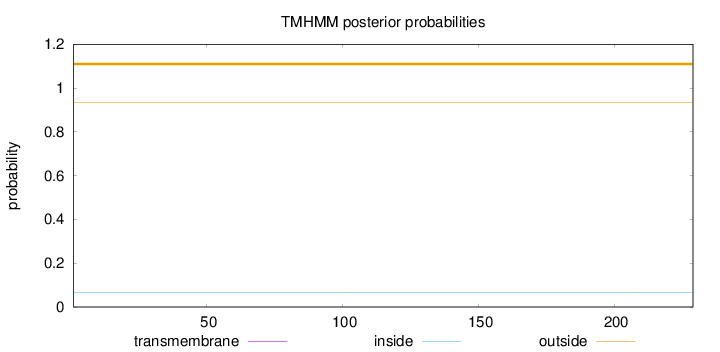
Topology
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0011
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.06669
outside
1 - 229
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
