Gene
KWMTBOMO16770
Pre Gene Modal
BGIBMGA009767
Annotation
PREDICTED:_phosphatidylinositol_4-phosphate_5-kinase_type-1_gamma_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.136
Sequence
CDS
ATGCTTCTACTGTGTTCCAGGCATCCAGCACGCTATAGGAGGCCTAGCGTCGAAGCCCGAACGCGATCTACTCATGCAGGACTTCATGACCGTCGAGACGACGAATTTCCCCCACGAGGGATCGAACCACACGCCCGCCCATCACTATTCCGAGTTCAATTCAAAACGTACGCGCCGATCGCGTTCCGTTACTTCAGAGATTTGTTTGGGATACAGCCGGACGATTTTTTGATGTCAATGTGCAGCGCACCACTCCGCGAGCTCAGCAACCCTGGCGCTTCAGGCAGCATCTTCTATCTGACGAACGACGATGAGTTCATCATAAAGACGGTACAGCACAAGGAGGGGGAGTTTCTGCAGAAACTACTGCCTGGATACTACTTGAACCTCGATCAGAACCCCCGCACGTTATTACCAAAGTTCTTCGGCCTCTATTGCTATCAGTGTAACAGCAAAAACGTGCGTCTGGTCGCCATGAACAATCTGTTGCCTTCGTCGATCAAAATGCACCAGAAATACGACCTCAAAGGGTCCACGTACAAACGTAAGGCAAGCAAAACAGAACGACAGAAAACCTCACCGACCTACAAGGATCTCGACTTCATGGAGCATCACAACGAGGGGATATTTCTGGAAGCGGACACGTACACGGCTTTGATAAAAACGATGCAGCGTGATTGTCGTGTATTGGAGAGCTTCAAGATCATGGACTACTCCCTCCTGGTCGGCATACATAACTTAGATGACGCCCAGCGCGAGAAGAACGAAGCCAGGAAACTGACGGAATCCGGTCAGAGCGATGGTGAGGATAACGAAGGGGAGACCAAGAGAACTGGACTCAATCGAACGAGATCCGAAGCCAGGGAGGATTTCGGGATCCAAGTTAAAAGGACACAATCGATAAACCGACAGCGTCTGGTCGCGCACTCGACAGCCATGGAGAGCATACAGGCGGAAAGCGAGCCCATAGATGAGGAGGAAGATGTCCCGTCCGGCGGCATCCCGGCCCGGAACGCGCGTGGCGAGCGTCTGCTGCTGTTCCTGGGCATCATCGACATACTCCAGTCGTATCGTCTCCGGAAGAAGCTCGAGCACACTTGGAAGAGCATGATACACGATGGCGACACTGTATCGGTCCACAGACCCAGCTTCTACGCTCAGAGATTCTTAGATTTCATGGCTAAAACCGTTTTCAAGAAGATACCTTCGTTGGACCTGCCGGAAATCAAGGGCAACCACAGGAAGTTCCGAACCCTCGTCACTAGTTACATAGCCCTGAAACACTCTCCTTCGAAGCGGAAGAGCATCGCGAAGCCGTCGCGGCCACAAGAGGAAATCGATACGCCAGGACGCTTCATCGGGCGACCTATCAAAGATTACAATCCGGAATATCTGAGACAGGCCCGACTGTCGCCCAGGTCTTCGGTACGTTCTAGCATGAGCGATTTTACCGATACGACCATATCGACGTGGAACCAGCGTCACCCGCCCGCGCCCGCCGAATACCCCTCGGTCCTTTCCGACAGACTGAAAATACTAGACAGGGACAGAATCAATAACTACCAGAGGCGTATCGATTTCGACACCTCGAACTTGACGGCGCGCGATCCGGGACCCTCGTCGGCATTTTACGCCGACAACGCGTACCGGTCCAACGTTCAGGAACGGATGGAGACGCTCTCGAACAGCTGA
Protein
MLLLCSRHPARYRRPSVEARTRSTHAGLHDRRDDEFPPRGIEPHARPSLFRVQFKTYAPIAFRYFRDLFGIQPDDFLMSMCSAPLRELSNPGASGSIFYLTNDDEFIIKTVQHKEGEFLQKLLPGYYLNLDQNPRTLLPKFFGLYCYQCNSKNVRLVAMNNLLPSSIKMHQKYDLKGSTYKRKASKTERQKTSPTYKDLDFMEHHNEGIFLEADTYTALIKTMQRDCRVLESFKIMDYSLLVGIHNLDDAQREKNEARKLTESGQSDGEDNEGETKRTGLNRTRSEAREDFGIQVKRTQSINRQRLVAHSTAMESIQAESEPIDEEEDVPSGGIPARNARGERLLLFLGIIDILQSYRLRKKLEHTWKSMIHDGDTVSVHRPSFYAQRFLDFMAKTVFKKIPSLDLPEIKGNHRKFRTLVTSYIALKHSPSKRKSIAKPSRPQEEIDTPGRFIGRPIKDYNPEYLRQARLSPRSSVRSSMSDFTDTTISTWNQRHPPAPAEYPSVLSDRLKILDRDRINNYQRRIDFDTSNLTARDPGPSSAFYADNAYRSNVQERMETLSNS
Summary
Uniprot
A0A194QZN8
A0A2H1VA20
A0A3S2LXW5
A0A151JRA7
E2A2X3
A0A2A4JJ66
+ More
F4W5I7 A0A0L7RBR5 A0A2J7Q593 A0A3L8DB04 A0A182R6J4 W5JFQ6 A0A1J1IIV5 A0A1J1IM69 A0A088AUC9 A0A0K8TLL4 A0A336MHI7 A0A023F342 T1HHL0 A0A0K8T3A5 A0A0K8T228 A0A336MD91 A0A146LV76 A0A195ERZ8 A0A151X330 A0A0A1X640 A0A0K8T8L7 A0A0J9RJA9 A0A0J9RJ52 A0A0K8T871 A0A0J9RIQ4 A0A0J9U8T3 A0A0J9RKB3 A0A0J9RJ45 B4MJL7 A0A0L0CBQ3 A0A0J9RIR0 A0A1I8MM18 A0A0N8P0F0 A0A0P8XQV7 Q0E8Y1 N6W6S8 A0A0B4K7E3 E2BHP5 Q9W1Y3 A0A0R1DZ25 A0A0Q5W2Q1 A0A3B0J4V6 A0A0R1DTK3 A0A1I8PWJ8 A0A0P9C078 A0A0Q5W035 A0A0Q5VNV1 A0A0Q5VP02 A0A0R1DT87 A0A0R1DYN1 A0A0C9RPZ2 A0A1I8PWI2 A0A0P9AGI9 B4LMW7 A0A0K8VSR8 A0A0R1DV98 A0A0Q5VNI7 A0A0K8U8S7 B5E1F8 A0A034VQ19 A0A0Q9WHA8 A0A0P8ZR90 W8BCY7 N6WCX8 A0A2J7Q590 A0A2J7Q592 A0A1W4VDB1 A0A1W4VQB2 A0A1W4VDK7 A0A1W4VRM7 A0A2A3EJ11 A0A0M4E9M6 W8BCZ2 A0A1Y1MVB0 A0A0R1DT90 A0A2J7Q587 A0A2J7Q595 A0A2J7Q599 A0A0R1DTJ0 A0A0R1DX03 A0A0Q5VXU6
F4W5I7 A0A0L7RBR5 A0A2J7Q593 A0A3L8DB04 A0A182R6J4 W5JFQ6 A0A1J1IIV5 A0A1J1IM69 A0A088AUC9 A0A0K8TLL4 A0A336MHI7 A0A023F342 T1HHL0 A0A0K8T3A5 A0A0K8T228 A0A336MD91 A0A146LV76 A0A195ERZ8 A0A151X330 A0A0A1X640 A0A0K8T8L7 A0A0J9RJA9 A0A0J9RJ52 A0A0K8T871 A0A0J9RIQ4 A0A0J9U8T3 A0A0J9RKB3 A0A0J9RJ45 B4MJL7 A0A0L0CBQ3 A0A0J9RIR0 A0A1I8MM18 A0A0N8P0F0 A0A0P8XQV7 Q0E8Y1 N6W6S8 A0A0B4K7E3 E2BHP5 Q9W1Y3 A0A0R1DZ25 A0A0Q5W2Q1 A0A3B0J4V6 A0A0R1DTK3 A0A1I8PWJ8 A0A0P9C078 A0A0Q5W035 A0A0Q5VNV1 A0A0Q5VP02 A0A0R1DT87 A0A0R1DYN1 A0A0C9RPZ2 A0A1I8PWI2 A0A0P9AGI9 B4LMW7 A0A0K8VSR8 A0A0R1DV98 A0A0Q5VNI7 A0A0K8U8S7 B5E1F8 A0A034VQ19 A0A0Q9WHA8 A0A0P8ZR90 W8BCY7 N6WCX8 A0A2J7Q590 A0A2J7Q592 A0A1W4VDB1 A0A1W4VQB2 A0A1W4VDK7 A0A1W4VRM7 A0A2A3EJ11 A0A0M4E9M6 W8BCZ2 A0A1Y1MVB0 A0A0R1DT90 A0A2J7Q587 A0A2J7Q595 A0A2J7Q599 A0A0R1DTJ0 A0A0R1DX03 A0A0Q5VXU6
Pubmed
EMBL
KQ460890
KPJ11008.1
ODYU01001460
SOQ37685.1
RSAL01000127
RVE46519.1
+ More
KQ978642 KYN29517.1 GL436229 EFN72215.1 NWSH01001293 PCG71816.1 GL887655 EGI70484.1 KQ414617 KOC68367.1 NEVH01017587 PNF23746.1 QOIP01000010 RLU17516.1 ADMH02001355 ETN62891.1 CVRI01000054 CRL00175.1 CRL00174.1 GDAI01002356 JAI15247.1 UFQT01000988 SSX28303.1 GBBI01003049 JAC15663.1 ACPB03000431 ACPB03000432 GBRD01016232 GBRD01006146 JAG59675.1 GBRD01016231 GBRD01006253 JAG59568.1 SSX28304.1 GDHC01007644 JAQ10985.1 KQ981993 KYN31003.1 KQ982562 KYQ54831.1 GBXI01007986 JAD06306.1 GBRD01004080 GBRD01004077 JAG61741.1 CM002911 KMY95897.1 KMY95901.1 GBRD01004078 GBRD01004075 JAG61743.1 KMY95898.1 KMY95900.1 KMY95899.1 KMY95896.1 CH963846 EDW72306.2 JRES01000644 KNC29670.1 KMY95903.1 CH902619 KPU76940.1 KPU76941.1 AE013599 ABI31111.3 CM000071 ENO01938.2 AFH08224.1 GL448322 EFN84793.1 AAF46919.3 CM000158 KRK00420.1 CH954179 KQS63000.1 OUUW01000001 SPP74643.1 KRK00421.1 KPU76944.1 KQS63006.1 KQS62999.1 KQS63011.1 KRK00411.1 KRK00417.1 GBYB01010570 JAG80337.1 KPU76939.1 CH940648 EDW62082.2 GDHF01024241 GDHF01010699 JAI28073.1 JAI41615.1 KRK00414.1 KQS63008.1 GDHF01029250 JAI23064.1 EDY69499.2 GAKP01015032 JAC43920.1 KRF80430.1 KPU76945.1 GAMC01015559 GAMC01015555 JAB90996.1 ENO01939.2 PNF23751.1 PNF23753.1 KZ288247 PBC31176.1 CP012524 ALC41646.1 GAMC01015557 GAMC01015554 JAB91001.1 GEZM01019595 JAV89601.1 KRK00416.1 PNF23748.1 PNF23750.1 PNF23752.1 KRK00413.1 KRK00419.1 KQS63001.1
KQ978642 KYN29517.1 GL436229 EFN72215.1 NWSH01001293 PCG71816.1 GL887655 EGI70484.1 KQ414617 KOC68367.1 NEVH01017587 PNF23746.1 QOIP01000010 RLU17516.1 ADMH02001355 ETN62891.1 CVRI01000054 CRL00175.1 CRL00174.1 GDAI01002356 JAI15247.1 UFQT01000988 SSX28303.1 GBBI01003049 JAC15663.1 ACPB03000431 ACPB03000432 GBRD01016232 GBRD01006146 JAG59675.1 GBRD01016231 GBRD01006253 JAG59568.1 SSX28304.1 GDHC01007644 JAQ10985.1 KQ981993 KYN31003.1 KQ982562 KYQ54831.1 GBXI01007986 JAD06306.1 GBRD01004080 GBRD01004077 JAG61741.1 CM002911 KMY95897.1 KMY95901.1 GBRD01004078 GBRD01004075 JAG61743.1 KMY95898.1 KMY95900.1 KMY95899.1 KMY95896.1 CH963846 EDW72306.2 JRES01000644 KNC29670.1 KMY95903.1 CH902619 KPU76940.1 KPU76941.1 AE013599 ABI31111.3 CM000071 ENO01938.2 AFH08224.1 GL448322 EFN84793.1 AAF46919.3 CM000158 KRK00420.1 CH954179 KQS63000.1 OUUW01000001 SPP74643.1 KRK00421.1 KPU76944.1 KQS63006.1 KQS62999.1 KQS63011.1 KRK00411.1 KRK00417.1 GBYB01010570 JAG80337.1 KPU76939.1 CH940648 EDW62082.2 GDHF01024241 GDHF01010699 JAI28073.1 JAI41615.1 KRK00414.1 KQS63008.1 GDHF01029250 JAI23064.1 EDY69499.2 GAKP01015032 JAC43920.1 KRF80430.1 KPU76945.1 GAMC01015559 GAMC01015555 JAB90996.1 ENO01939.2 PNF23751.1 PNF23753.1 KZ288247 PBC31176.1 CP012524 ALC41646.1 GAMC01015557 GAMC01015554 JAB91001.1 GEZM01019595 JAV89601.1 KRK00416.1 PNF23748.1 PNF23750.1 PNF23752.1 KRK00413.1 KRK00419.1 KQS63001.1
Proteomes
UP000053240
UP000283053
UP000078492
UP000000311
UP000218220
UP000007755
+ More
UP000053825 UP000235965 UP000279307 UP000075900 UP000000673 UP000183832 UP000005203 UP000015103 UP000078541 UP000075809 UP000007798 UP000037069 UP000095301 UP000007801 UP000000803 UP000001819 UP000008237 UP000002282 UP000008711 UP000268350 UP000095300 UP000008792 UP000192221 UP000242457 UP000092553
UP000053825 UP000235965 UP000279307 UP000075900 UP000000673 UP000183832 UP000005203 UP000015103 UP000078541 UP000075809 UP000007798 UP000037069 UP000095301 UP000007801 UP000000803 UP000001819 UP000008237 UP000002282 UP000008711 UP000268350 UP000095300 UP000008792 UP000192221 UP000242457 UP000092553
Pfam
PF01504 PIP5K
Interpro
Gene 3D
ProteinModelPortal
A0A194QZN8
A0A2H1VA20
A0A3S2LXW5
A0A151JRA7
E2A2X3
A0A2A4JJ66
+ More
F4W5I7 A0A0L7RBR5 A0A2J7Q593 A0A3L8DB04 A0A182R6J4 W5JFQ6 A0A1J1IIV5 A0A1J1IM69 A0A088AUC9 A0A0K8TLL4 A0A336MHI7 A0A023F342 T1HHL0 A0A0K8T3A5 A0A0K8T228 A0A336MD91 A0A146LV76 A0A195ERZ8 A0A151X330 A0A0A1X640 A0A0K8T8L7 A0A0J9RJA9 A0A0J9RJ52 A0A0K8T871 A0A0J9RIQ4 A0A0J9U8T3 A0A0J9RKB3 A0A0J9RJ45 B4MJL7 A0A0L0CBQ3 A0A0J9RIR0 A0A1I8MM18 A0A0N8P0F0 A0A0P8XQV7 Q0E8Y1 N6W6S8 A0A0B4K7E3 E2BHP5 Q9W1Y3 A0A0R1DZ25 A0A0Q5W2Q1 A0A3B0J4V6 A0A0R1DTK3 A0A1I8PWJ8 A0A0P9C078 A0A0Q5W035 A0A0Q5VNV1 A0A0Q5VP02 A0A0R1DT87 A0A0R1DYN1 A0A0C9RPZ2 A0A1I8PWI2 A0A0P9AGI9 B4LMW7 A0A0K8VSR8 A0A0R1DV98 A0A0Q5VNI7 A0A0K8U8S7 B5E1F8 A0A034VQ19 A0A0Q9WHA8 A0A0P8ZR90 W8BCY7 N6WCX8 A0A2J7Q590 A0A2J7Q592 A0A1W4VDB1 A0A1W4VQB2 A0A1W4VDK7 A0A1W4VRM7 A0A2A3EJ11 A0A0M4E9M6 W8BCZ2 A0A1Y1MVB0 A0A0R1DT90 A0A2J7Q587 A0A2J7Q595 A0A2J7Q599 A0A0R1DTJ0 A0A0R1DX03 A0A0Q5VXU6
F4W5I7 A0A0L7RBR5 A0A2J7Q593 A0A3L8DB04 A0A182R6J4 W5JFQ6 A0A1J1IIV5 A0A1J1IM69 A0A088AUC9 A0A0K8TLL4 A0A336MHI7 A0A023F342 T1HHL0 A0A0K8T3A5 A0A0K8T228 A0A336MD91 A0A146LV76 A0A195ERZ8 A0A151X330 A0A0A1X640 A0A0K8T8L7 A0A0J9RJA9 A0A0J9RJ52 A0A0K8T871 A0A0J9RIQ4 A0A0J9U8T3 A0A0J9RKB3 A0A0J9RJ45 B4MJL7 A0A0L0CBQ3 A0A0J9RIR0 A0A1I8MM18 A0A0N8P0F0 A0A0P8XQV7 Q0E8Y1 N6W6S8 A0A0B4K7E3 E2BHP5 Q9W1Y3 A0A0R1DZ25 A0A0Q5W2Q1 A0A3B0J4V6 A0A0R1DTK3 A0A1I8PWJ8 A0A0P9C078 A0A0Q5W035 A0A0Q5VNV1 A0A0Q5VP02 A0A0R1DT87 A0A0R1DYN1 A0A0C9RPZ2 A0A1I8PWI2 A0A0P9AGI9 B4LMW7 A0A0K8VSR8 A0A0R1DV98 A0A0Q5VNI7 A0A0K8U8S7 B5E1F8 A0A034VQ19 A0A0Q9WHA8 A0A0P8ZR90 W8BCY7 N6WCX8 A0A2J7Q590 A0A2J7Q592 A0A1W4VDB1 A0A1W4VQB2 A0A1W4VDK7 A0A1W4VRM7 A0A2A3EJ11 A0A0M4E9M6 W8BCZ2 A0A1Y1MVB0 A0A0R1DT90 A0A2J7Q587 A0A2J7Q595 A0A2J7Q599 A0A0R1DTJ0 A0A0R1DX03 A0A0Q5VXU6
PDB
5E3U
E-value=5.27845e-115,
Score=1061
Ontologies
GO
PANTHER
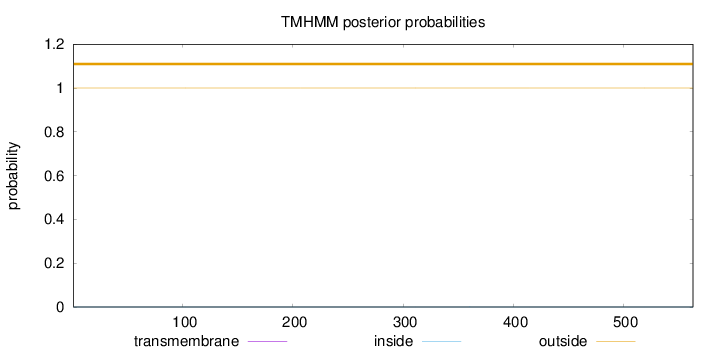
Topology
Length:
563
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00193
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.00031
outside
1 - 563
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
