Pre Gene Modal
BGIBMGA000232
Annotation
calcyphosphine_isoform_2_[Bombyx_mori]
Full name
Anoctamin
Location in the cell
Cytoplasmic Reliability : 1.68 Nuclear Reliability : 2.283
Sequence
CDS
ATGTATTTAGGAAAACCGAAAACATCTTCCAACATTGCGTGGCACAGACCGATGTCTGCTGGTTCCGTCCAGGAGGAAGAGCTGATGCAAAAGAGTGCTCGGATCTTGGGACTTGGAAGAATTTTCCGTCGCATGGATGATGATGGAAGTAAACTACTGAACAAAGAAGAATTCCTTTATGGCATCAAGGAAACAGGGCTGGAACTTAATAAAAGTGAAGCCGAAGAACTCTTCAGTCAATTCGACACAGACAGTAGTGGCTCAATCAGTCTTGATGAATTCCTTATTAAAATCCGTCCTCCCATGTCGGAATCGCGTCGTAACATTGTAGAACAAGCATTCAAGAAGCTTGACAAGACTGGTGACGGTGCAATCACAATTGATGACATTAAAGGTGTTTACTCTGTCGATTCTCAGACCAGATATAAGAGCGGCGAAGAAACTGAAGATGTAATAATGAAGAGGTTCCTCGCGAATTTTGAGTCCGATGGAACAGTCGATGGGAAGGTTACTCTTGAAGAGTTCATGAACTACTACAGTGGCATCAGTGTCTCCATTGACAACGACTGCTATTTTGACCTAATGATGCGTCAGGCGTACAAGCTCTAA
Protein
MYLGKPKTSSNIAWHRPMSAGSVQEEELMQKSARILGLGRIFRRMDDDGSKLLNKEEFLYGIKETGLELNKSEAEELFSQFDTDSSGSISLDEFLIKIRPPMSESRRNIVEQAFKKLDKTGDGAITIDDIKGVYSVDSQTRYKSGEETEDVIMKRFLANFESDGTVDGKVTLEEFMNYYSGISVSIDNDCYFDLMMRQAYKL
Summary
Similarity
Belongs to the anoctamin family.
Uniprot
Q1HQ30
Q1HQ31
H9ISK5
A0A2A4J6U1
A0A2H1VL40
Q9U506
+ More
A0A3B0ENA0 I4DKM0 S4PBQ0 A0A3S2PIN7 A0A212F194 A0A0N1IE99 A0A194QDN5 A0A023EM24 D6WJF8 T1DIN5 Q1HQR5 A0A182KRY2 A0A2C9GPR9 A0A084VF79 A0A182XZF7 A0A2M4CYL6 W5JS58 A0A182JW96 A0A2M4BZE4 A0A1B0CR65 A0A182GRE8 A0A182P9X3 A0A182X6H7 Q7PTC6 A0A182V2I9 A0A182MUJ3 A0A2M4A9S2 A0A2M4BZI5 A0A182FGP9 A0A182NHG0 A0A2M3Z0H1 A0A182U749 A0A182RB53 A0A2M4BZC8 A0A182QN21 A0A2P8Z882 A0A1Q3FQX0 A0A182J996 U5EW81 A0A067R7H7 F4WWD8 B0VZS4 A0A069DQP9 A0A2J7PQZ4 A0A0K8SAT4 A0A0A9Y698 A0A154PTP8 A0A0K8SAG8 A0A182GAK1 A0A146LG55 A0A151WRR5 A0A0N0BJG3 A0A0A9Y4Q9 E2AMV5 T1E1R2 A0A0V0GA11 A0A151IJS0 A0A023EZ00 A0A087ZSL7 A0A2A3EEL4 A0A182TT88 A0A067R9C2 A0A151JBD8 T1I0D3 A0A182WIV0 A0A195BNK8 A0A158NKK9 A0A336M867 A0A336MB06 A0A1J1HXR1 E2C9D1 A0A195FCN4 A0A0L7QST0 A0A1L8DMW8 U4U1K0 A0A1Y1LBX1 A0A182SJQ0 J3JXD5 K7J4H7 A0A232F085 A0A1D2MHN8 A0A1B6C2W5 A0A161MSS7 A0A026WU42 A0A1B6GXM9 A0A0C9QP10 A0A1B6K4W5 A0A1B6M9I8 A0A1B6MFG5 E9IFE1 A0A336KZL7 A0A2P8Z7B5 A0A2J7PSD0 E0VRQ9
A0A3B0ENA0 I4DKM0 S4PBQ0 A0A3S2PIN7 A0A212F194 A0A0N1IE99 A0A194QDN5 A0A023EM24 D6WJF8 T1DIN5 Q1HQR5 A0A182KRY2 A0A2C9GPR9 A0A084VF79 A0A182XZF7 A0A2M4CYL6 W5JS58 A0A182JW96 A0A2M4BZE4 A0A1B0CR65 A0A182GRE8 A0A182P9X3 A0A182X6H7 Q7PTC6 A0A182V2I9 A0A182MUJ3 A0A2M4A9S2 A0A2M4BZI5 A0A182FGP9 A0A182NHG0 A0A2M3Z0H1 A0A182U749 A0A182RB53 A0A2M4BZC8 A0A182QN21 A0A2P8Z882 A0A1Q3FQX0 A0A182J996 U5EW81 A0A067R7H7 F4WWD8 B0VZS4 A0A069DQP9 A0A2J7PQZ4 A0A0K8SAT4 A0A0A9Y698 A0A154PTP8 A0A0K8SAG8 A0A182GAK1 A0A146LG55 A0A151WRR5 A0A0N0BJG3 A0A0A9Y4Q9 E2AMV5 T1E1R2 A0A0V0GA11 A0A151IJS0 A0A023EZ00 A0A087ZSL7 A0A2A3EEL4 A0A182TT88 A0A067R9C2 A0A151JBD8 T1I0D3 A0A182WIV0 A0A195BNK8 A0A158NKK9 A0A336M867 A0A336MB06 A0A1J1HXR1 E2C9D1 A0A195FCN4 A0A0L7QST0 A0A1L8DMW8 U4U1K0 A0A1Y1LBX1 A0A182SJQ0 J3JXD5 K7J4H7 A0A232F085 A0A1D2MHN8 A0A1B6C2W5 A0A161MSS7 A0A026WU42 A0A1B6GXM9 A0A0C9QP10 A0A1B6K4W5 A0A1B6M9I8 A0A1B6MFG5 E9IFE1 A0A336KZL7 A0A2P8Z7B5 A0A2J7PSD0 E0VRQ9
Pubmed
19121390
10620045
22651552
23622113
22118469
26354079
+ More
24945155 18362917 19820115 24330624 17204158 17510324 20966253 24438588 25244985 20920257 23761445 26483478 12364791 14747013 17210077 29403074 24845553 21719571 26334808 26823975 25401762 20798317 25474469 21347285 23537049 28004739 22516182 20075255 28648823 27289101 24508170 30249741 21282665 20566863
24945155 18362917 19820115 24330624 17204158 17510324 20966253 24438588 25244985 20920257 23761445 26483478 12364791 14747013 17210077 29403074 24845553 21719571 26334808 26823975 25401762 20798317 25474469 21347285 23537049 28004739 22516182 20075255 28648823 27289101 24508170 30249741 21282665 20566863
EMBL
DQ443222
ABF51311.1
DQ443221
ABF51310.1
BABH01013495
NWSH01002941
+ More
PCG67244.1 ODYU01003154 SOQ41550.1 AF117582 AAF16704.1 RBVL01000197 RKO10832.1 AK401838 BAM18460.1 GAIX01004506 JAA88054.1 RSAL01000021 RVE52503.1 AGBW02010923 OWR47509.1 KQ460968 KPJ10222.1 KQ459324 KPJ01566.1 GAPW01004254 JAC09344.1 KQ971342 EFA03890.2 GALA01000851 JAA94001.1 DQ440379 CH477513 ABF18412.1 EAT39728.1 APCN01000220 ATLV01012365 KE524785 KFB36623.1 GGFL01006209 MBW70387.1 ADMH02000582 ETN65760.1 GGFJ01009282 MBW58423.1 AJWK01024422 JXUM01082355 KQ563307 KXJ74089.1 AAAB01008807 EAA04113.4 AXCM01000560 GGFK01004235 MBW37556.1 GGFJ01009283 MBW58424.1 GGFM01001269 MBW22020.1 GGFJ01009285 MBW58426.1 AXCN02000591 PYGN01000151 PSN52705.1 GFDL01005024 JAV30021.1 GANO01000681 JAB59190.1 KK852651 KDR19448.1 GL888406 EGI61480.1 DS231815 EDS35166.1 GBGD01002932 JAC85957.1 NEVH01022634 PNF18755.1 GBRD01015557 GDHC01015624 GDHC01014709 JAG50269.1 JAQ03005.1 JAQ03920.1 GBHO01017011 JAG26593.1 KQ435090 KZC14570.1 GBRD01015555 JAG50271.1 JXUM01008681 KQ560266 KXJ83465.1 GDHC01012653 JAQ05976.1 KQ982805 KYQ50497.1 KQ435718 KOX78972.1 GBHO01017008 GBRD01015556 GDHC01005823 JAG26596.1 JAG50270.1 JAQ12806.1 GL440936 EFN65229.1 GALA01001424 JAA93428.1 GECL01001233 JAP04891.1 KQ977294 KYN03882.1 GBBI01004783 JAC13929.1 KZ288266 PBC30195.1 KK852611 KDR20276.1 KQ979182 KYN22309.1 ACPB03005879 KQ976439 KYM86781.1 ADTU01018872 UFQT01000679 SSX26512.1 SSX26511.1 CVRI01000035 CRK92872.1 GL453806 EFN75476.1 KQ981693 KYN37804.1 KQ414756 KOC61609.1 GFDF01006288 JAV07796.1 KB630812 KB630813 ERL83880.1 ERL83885.1 GEZM01063622 JAV69046.1 BT127904 AEE62866.1 NNAY01001377 OXU24176.1 LJIJ01001230 ODM92441.1 GEDC01029445 GEDC01002030 JAS07853.1 JAS35268.1 GEMB01000624 JAS02509.1 KK107105 QOIP01000011 EZA59547.1 RLU17223.1 GECZ01002591 JAS67178.1 GBYB01005289 JAG75056.1 GECU01001207 JAT06500.1 GEBQ01007390 JAT32587.1 GEBQ01005280 JAT34697.1 GL762838 EFZ20689.1 UFQS01001307 UFQT01001307 SSX10237.1 SSX29925.1 PYGN01000164 PSN52383.1 NEVH01021933 PNF19230.1 DS235494 EEB16065.1
PCG67244.1 ODYU01003154 SOQ41550.1 AF117582 AAF16704.1 RBVL01000197 RKO10832.1 AK401838 BAM18460.1 GAIX01004506 JAA88054.1 RSAL01000021 RVE52503.1 AGBW02010923 OWR47509.1 KQ460968 KPJ10222.1 KQ459324 KPJ01566.1 GAPW01004254 JAC09344.1 KQ971342 EFA03890.2 GALA01000851 JAA94001.1 DQ440379 CH477513 ABF18412.1 EAT39728.1 APCN01000220 ATLV01012365 KE524785 KFB36623.1 GGFL01006209 MBW70387.1 ADMH02000582 ETN65760.1 GGFJ01009282 MBW58423.1 AJWK01024422 JXUM01082355 KQ563307 KXJ74089.1 AAAB01008807 EAA04113.4 AXCM01000560 GGFK01004235 MBW37556.1 GGFJ01009283 MBW58424.1 GGFM01001269 MBW22020.1 GGFJ01009285 MBW58426.1 AXCN02000591 PYGN01000151 PSN52705.1 GFDL01005024 JAV30021.1 GANO01000681 JAB59190.1 KK852651 KDR19448.1 GL888406 EGI61480.1 DS231815 EDS35166.1 GBGD01002932 JAC85957.1 NEVH01022634 PNF18755.1 GBRD01015557 GDHC01015624 GDHC01014709 JAG50269.1 JAQ03005.1 JAQ03920.1 GBHO01017011 JAG26593.1 KQ435090 KZC14570.1 GBRD01015555 JAG50271.1 JXUM01008681 KQ560266 KXJ83465.1 GDHC01012653 JAQ05976.1 KQ982805 KYQ50497.1 KQ435718 KOX78972.1 GBHO01017008 GBRD01015556 GDHC01005823 JAG26596.1 JAG50270.1 JAQ12806.1 GL440936 EFN65229.1 GALA01001424 JAA93428.1 GECL01001233 JAP04891.1 KQ977294 KYN03882.1 GBBI01004783 JAC13929.1 KZ288266 PBC30195.1 KK852611 KDR20276.1 KQ979182 KYN22309.1 ACPB03005879 KQ976439 KYM86781.1 ADTU01018872 UFQT01000679 SSX26512.1 SSX26511.1 CVRI01000035 CRK92872.1 GL453806 EFN75476.1 KQ981693 KYN37804.1 KQ414756 KOC61609.1 GFDF01006288 JAV07796.1 KB630812 KB630813 ERL83880.1 ERL83885.1 GEZM01063622 JAV69046.1 BT127904 AEE62866.1 NNAY01001377 OXU24176.1 LJIJ01001230 ODM92441.1 GEDC01029445 GEDC01002030 JAS07853.1 JAS35268.1 GEMB01000624 JAS02509.1 KK107105 QOIP01000011 EZA59547.1 RLU17223.1 GECZ01002591 JAS67178.1 GBYB01005289 JAG75056.1 GECU01001207 JAT06500.1 GEBQ01007390 JAT32587.1 GEBQ01005280 JAT34697.1 GL762838 EFZ20689.1 UFQS01001307 UFQT01001307 SSX10237.1 SSX29925.1 PYGN01000164 PSN52383.1 NEVH01021933 PNF19230.1 DS235494 EEB16065.1
Proteomes
UP000005204
UP000218220
UP000276569
UP000283053
UP000007151
UP000053240
+ More
UP000053268 UP000007266 UP000008820 UP000075882 UP000075840 UP000030765 UP000076408 UP000000673 UP000075881 UP000092461 UP000069940 UP000249989 UP000075885 UP000076407 UP000007062 UP000075903 UP000075883 UP000069272 UP000075884 UP000075902 UP000075900 UP000075886 UP000245037 UP000075880 UP000027135 UP000007755 UP000002320 UP000235965 UP000076502 UP000075809 UP000053105 UP000000311 UP000078542 UP000005203 UP000242457 UP000078492 UP000015103 UP000075920 UP000078540 UP000005205 UP000183832 UP000008237 UP000078541 UP000053825 UP000030742 UP000075901 UP000002358 UP000215335 UP000094527 UP000053097 UP000279307 UP000009046
UP000053268 UP000007266 UP000008820 UP000075882 UP000075840 UP000030765 UP000076408 UP000000673 UP000075881 UP000092461 UP000069940 UP000249989 UP000075885 UP000076407 UP000007062 UP000075903 UP000075883 UP000069272 UP000075884 UP000075902 UP000075900 UP000075886 UP000245037 UP000075880 UP000027135 UP000007755 UP000002320 UP000235965 UP000076502 UP000075809 UP000053105 UP000000311 UP000078542 UP000005203 UP000242457 UP000078492 UP000015103 UP000075920 UP000078540 UP000005205 UP000183832 UP000008237 UP000078541 UP000053825 UP000030742 UP000075901 UP000002358 UP000215335 UP000094527 UP000053097 UP000279307 UP000009046
Pfam
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
Q1HQ30
Q1HQ31
H9ISK5
A0A2A4J6U1
A0A2H1VL40
Q9U506
+ More
A0A3B0ENA0 I4DKM0 S4PBQ0 A0A3S2PIN7 A0A212F194 A0A0N1IE99 A0A194QDN5 A0A023EM24 D6WJF8 T1DIN5 Q1HQR5 A0A182KRY2 A0A2C9GPR9 A0A084VF79 A0A182XZF7 A0A2M4CYL6 W5JS58 A0A182JW96 A0A2M4BZE4 A0A1B0CR65 A0A182GRE8 A0A182P9X3 A0A182X6H7 Q7PTC6 A0A182V2I9 A0A182MUJ3 A0A2M4A9S2 A0A2M4BZI5 A0A182FGP9 A0A182NHG0 A0A2M3Z0H1 A0A182U749 A0A182RB53 A0A2M4BZC8 A0A182QN21 A0A2P8Z882 A0A1Q3FQX0 A0A182J996 U5EW81 A0A067R7H7 F4WWD8 B0VZS4 A0A069DQP9 A0A2J7PQZ4 A0A0K8SAT4 A0A0A9Y698 A0A154PTP8 A0A0K8SAG8 A0A182GAK1 A0A146LG55 A0A151WRR5 A0A0N0BJG3 A0A0A9Y4Q9 E2AMV5 T1E1R2 A0A0V0GA11 A0A151IJS0 A0A023EZ00 A0A087ZSL7 A0A2A3EEL4 A0A182TT88 A0A067R9C2 A0A151JBD8 T1I0D3 A0A182WIV0 A0A195BNK8 A0A158NKK9 A0A336M867 A0A336MB06 A0A1J1HXR1 E2C9D1 A0A195FCN4 A0A0L7QST0 A0A1L8DMW8 U4U1K0 A0A1Y1LBX1 A0A182SJQ0 J3JXD5 K7J4H7 A0A232F085 A0A1D2MHN8 A0A1B6C2W5 A0A161MSS7 A0A026WU42 A0A1B6GXM9 A0A0C9QP10 A0A1B6K4W5 A0A1B6M9I8 A0A1B6MFG5 E9IFE1 A0A336KZL7 A0A2P8Z7B5 A0A2J7PSD0 E0VRQ9
A0A3B0ENA0 I4DKM0 S4PBQ0 A0A3S2PIN7 A0A212F194 A0A0N1IE99 A0A194QDN5 A0A023EM24 D6WJF8 T1DIN5 Q1HQR5 A0A182KRY2 A0A2C9GPR9 A0A084VF79 A0A182XZF7 A0A2M4CYL6 W5JS58 A0A182JW96 A0A2M4BZE4 A0A1B0CR65 A0A182GRE8 A0A182P9X3 A0A182X6H7 Q7PTC6 A0A182V2I9 A0A182MUJ3 A0A2M4A9S2 A0A2M4BZI5 A0A182FGP9 A0A182NHG0 A0A2M3Z0H1 A0A182U749 A0A182RB53 A0A2M4BZC8 A0A182QN21 A0A2P8Z882 A0A1Q3FQX0 A0A182J996 U5EW81 A0A067R7H7 F4WWD8 B0VZS4 A0A069DQP9 A0A2J7PQZ4 A0A0K8SAT4 A0A0A9Y698 A0A154PTP8 A0A0K8SAG8 A0A182GAK1 A0A146LG55 A0A151WRR5 A0A0N0BJG3 A0A0A9Y4Q9 E2AMV5 T1E1R2 A0A0V0GA11 A0A151IJS0 A0A023EZ00 A0A087ZSL7 A0A2A3EEL4 A0A182TT88 A0A067R9C2 A0A151JBD8 T1I0D3 A0A182WIV0 A0A195BNK8 A0A158NKK9 A0A336M867 A0A336MB06 A0A1J1HXR1 E2C9D1 A0A195FCN4 A0A0L7QST0 A0A1L8DMW8 U4U1K0 A0A1Y1LBX1 A0A182SJQ0 J3JXD5 K7J4H7 A0A232F085 A0A1D2MHN8 A0A1B6C2W5 A0A161MSS7 A0A026WU42 A0A1B6GXM9 A0A0C9QP10 A0A1B6K4W5 A0A1B6M9I8 A0A1B6MFG5 E9IFE1 A0A336KZL7 A0A2P8Z7B5 A0A2J7PSD0 E0VRQ9
PDB
3E3R
E-value=1.79228e-39,
Score=405
Ontologies
GO
PANTHER
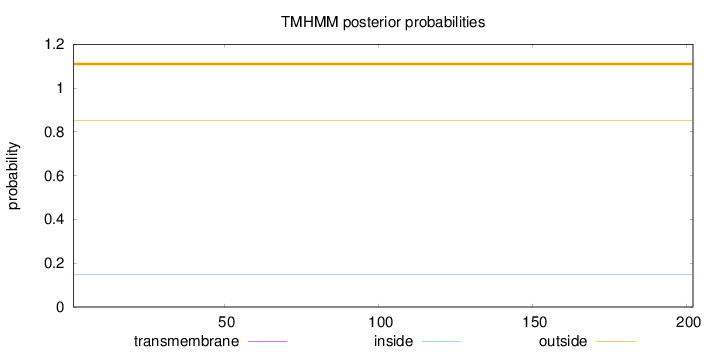
Topology
Subcellular location
Membrane
Length:
202
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00182
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14688
outside
1 - 202
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
