Gene
KWMTBOMO16757
Pre Gene Modal
BGIBMGA000932
Annotation
PREDICTED:_serine/threonine-protein_phosphatase_rdgC_[Bombyx_mori]
Full name
Serine/threonine-protein phosphatase with EF-hands
+ More
Serine/threonine-protein phosphatase
Serine/threonine-protein phosphatase rdgC
Serine/threonine-protein phosphatase
Serine/threonine-protein phosphatase rdgC
Alternative Name
Retinal degeneration C protein
Location in the cell
Cytoplasmic Reliability : 1.399 Mitochondrial Reliability : 1.289 Nuclear Reliability : 1.262
Sequence
CDS
ATGCGATATAAAGAGATTTACGTGTCGCTTCTGCGGCCGCCGGTGATCGAGGGCTCAACGGCCGCGGAGTCGATAGATAAGGTGGAGTGGAAGCAGGTGTTTGATATACTGTGGTCGGATCCACAATGCATCGGAGGATGTGTGGCAAATGCGCTTCGAGGAGCAGGAACCTACTTCGGCCCTGATGTAACAGCGTCGTCGTTCCTCCAGAAGAATAAGCTGAGCTTCATAATTAGAAGTCACGAGTGCAAGCCCGGTGGATACGAAATCATGCATGGCGGTAATGTAATTACAATTTTCTCGGCGTCGAACTATTACGAGTTGGGTTCCAACAAGGGCGCTTACCTGAAACTATGCGGGAACTCGTTGGAGCGCCAGTTCGTGCAATACACGGCGGCCGTGTCTCGCACGCGGCGTCTCACCTTCAGGCAGCGGGTTGGCCTTGTCGAATCGTCCGCCATGAGGGAGCTACACAATCAGATTATGATGGCCAGGAGATCTCTGGAAGCTACTTTCCGTAGCTTGGACTCGGAGCAAAGTGGTTTCATATCTATAAGCGATTGGTGTCAAGCCATGGAGGAGACAACACACCTCGGTCTCCCGTGGAGGATGCTTAAAGACAAGCTTGTTCGCACAGATCCCGAGACACGCAAAGTACGTTACATGGATACATTCGAACTCAACACCAATAGGAACGGACGAAAATCTGACACGCTGACAGTCGTCGAGACTTTGTATAAAAATCAAAGAAGCTTGGAGGCGATCTTCAAGATATTGGATAAAGATAATTCTGGTTACATAACGTTGGAAGAATTTTCCGAGGCTTGCCAGCTGATCGGCAAGTACATGCCGAATCCGATGACGCAGGAGCAGCTCGTCGACATCTGTCGGCTCATGGACATCAACAAGGACGGCCTGGTTGATCTCAACGAGTTCCTCGAGAGTTTCCGTCTCGCCGAAAAGAGTTTACACGAGCAGGAAAGGATGTACGAAGACATGCAGGCTACCTGA
Protein
MRYKEIYVSLLRPPVIEGSTAAESIDKVEWKQVFDILWSDPQCIGGCVANALRGAGTYFGPDVTASSFLQKNKLSFIIRSHECKPGGYEIMHGGNVITIFSASNYYELGSNKGAYLKLCGNSLERQFVQYTAAVSRTRRLTFRQRVGLVESSAMRELHNQIMMARRSLEATFRSLDSEQSGFISISDWCQAMEETTHLGLPWRMLKDKLVRTDPETRKVRYMDTFELNTNRNGRKSDTLTVVETLYKNQRSLEAIFKILDKDNSGYITLEEFSEACQLIGKYMPNPMTQEQLVDICRLMDINKDGLVDLNEFLESFRLAEKSLHEQERMYEDMQAT
Summary
Description
Phosphatase required to prevent light-induced retinal degeneration.
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
Cofactor
Mn(2+)
Similarity
Belongs to the PPP phosphatase family.
Keywords
Calcium
Complete proteome
Hydrolase
Magnesium
Manganese
Metal-binding
Protein phosphatase
Reference proteome
Repeat
Sensory transduction
Vision
Feature
chain Serine/threonine-protein phosphatase
Uniprot
H9IUK2
A0A194RLP1
A0A194PJL2
A0A437BJB7
A0A212EZK5
A0A2H1VHP4
+ More
A0A2A4JTJ8 A0A0L7L4I0 A0A0T6AY67 D2A162 A0A1W4WIT1 A0A1B6HRT7 A0A1B6LRB5 N6UVB8 Q7Q2Z9 A0A224XB96 Q17FS3 A0A182K8J4 A0A182WR55 A0A182WF17 A0A1S4F4H5 U4US14 A0A067R243 A0A182RFE2 E0VT76 A0A182FTV4 A0A182NPL6 A0A1S4H6J9 A0A453YJM5 A0A084WI84 A0A182PU84 A0A1B6DP80 W5JP23 A0A1B6C198 A0A336KR11 A0A0A9XJD6 A0A1B0CWS4 A0A0L0BQ75 T1I8U1 A0A1I8ML02 A0A1I8QDX9 N6TJK7 B4QRV8 A0A182V902 A0A182MN75 A0A182IJK1 A0A0J9UPV3 B3NE58 P40421 A0A0R1E3V8 B4IA68 A0A0Q5UKB6 A0A0J9RYJ5 A0A0R1E263 A0A336KZ48 A0A0J9RZW2 A0A0Q5U6J1 B4PFC5 A0A1W4VUR9 A0A182G6D2 B3M4R3 A0A0P8XTZ4 W8ANN1 A0A0P8XTK4 B4MKU9 A0A034WBT5 A0A3B0KHY3 A0A1A9VSD6 A0A0R3P7M8 A0A0R3P3H1 A0A1A9UQS4 A0A0R3P3E4 A0A3B0K9C5 A0A1A9Z5T7 A0A0R3P3C9 B5DPA9 B4GR08 A0A1A9WRH1 B4LG02 A0A0Q9WVD8 A0A1D2MFK2 A0A1B0G8C3 A0A0M4EZY5 A0A1A9Z4V7 B4KV71 A0A0Q9XQA2 B4IYR3 A0A1B0G581 A0A0Q9XDH4 A0A226E4L8 A0A2P2I617 A0A131XLU5 A0A224Z6H9 A0A131YUK5 A0A1E1XP01 A0A1B0B3L8 A0A1A9Z2Z5 A0A293LIW1 A0A147BQQ3
A0A2A4JTJ8 A0A0L7L4I0 A0A0T6AY67 D2A162 A0A1W4WIT1 A0A1B6HRT7 A0A1B6LRB5 N6UVB8 Q7Q2Z9 A0A224XB96 Q17FS3 A0A182K8J4 A0A182WR55 A0A182WF17 A0A1S4F4H5 U4US14 A0A067R243 A0A182RFE2 E0VT76 A0A182FTV4 A0A182NPL6 A0A1S4H6J9 A0A453YJM5 A0A084WI84 A0A182PU84 A0A1B6DP80 W5JP23 A0A1B6C198 A0A336KR11 A0A0A9XJD6 A0A1B0CWS4 A0A0L0BQ75 T1I8U1 A0A1I8ML02 A0A1I8QDX9 N6TJK7 B4QRV8 A0A182V902 A0A182MN75 A0A182IJK1 A0A0J9UPV3 B3NE58 P40421 A0A0R1E3V8 B4IA68 A0A0Q5UKB6 A0A0J9RYJ5 A0A0R1E263 A0A336KZ48 A0A0J9RZW2 A0A0Q5U6J1 B4PFC5 A0A1W4VUR9 A0A182G6D2 B3M4R3 A0A0P8XTZ4 W8ANN1 A0A0P8XTK4 B4MKU9 A0A034WBT5 A0A3B0KHY3 A0A1A9VSD6 A0A0R3P7M8 A0A0R3P3H1 A0A1A9UQS4 A0A0R3P3E4 A0A3B0K9C5 A0A1A9Z5T7 A0A0R3P3C9 B5DPA9 B4GR08 A0A1A9WRH1 B4LG02 A0A0Q9WVD8 A0A1D2MFK2 A0A1B0G8C3 A0A0M4EZY5 A0A1A9Z4V7 B4KV71 A0A0Q9XQA2 B4IYR3 A0A1B0G581 A0A0Q9XDH4 A0A226E4L8 A0A2P2I617 A0A131XLU5 A0A224Z6H9 A0A131YUK5 A0A1E1XP01 A0A1B0B3L8 A0A1A9Z2Z5 A0A293LIW1 A0A147BQQ3
EC Number
3.1.3.16
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 12364791 17510324 24845553 20566863 24438588 20920257 23761445 25401762 26823975 26108605 25315136 17994087 22936249 1316807 10731132 12537572 17550304 26483478 24495485 25348373 15632085 23185243 18057021 27289101 28049606 28797301 26830274 29209593 29652888
23537049 12364791 17510324 24845553 20566863 24438588 20920257 23761445 25401762 26823975 26108605 25315136 17994087 22936249 1316807 10731132 12537572 17550304 26483478 24495485 25348373 15632085 23185243 18057021 27289101 28049606 28797301 26830274 29209593 29652888
EMBL
BABH01000935
KQ460205
KPJ16901.1
KQ459606
KPI91275.1
RSAL01000047
+ More
RVE50532.1 AGBW02011268 OWR46920.1 ODYU01002624 SOQ40363.1 NWSH01000662 PCG74944.1 JTDY01003056 KOB70236.1 LJIG01022522 KRT80131.1 KQ971338 EFA01582.2 GECU01030324 JAS77382.1 GEBQ01013744 JAT26233.1 APGK01009376 APGK01009377 KB737932 ENN82832.1 AAAB01008966 EAA12933.4 GFTR01006749 JAW09677.1 CH477268 EAT45428.1 KB632337 ERL92855.1 KK852756 KDR17087.1 DS235760 EEB16582.1 APCN01001959 APCN01001960 ATLV01023923 KE525347 KFB49928.1 GEDC01009827 JAS27471.1 ADMH02000929 ETN64645.1 GEDC01030006 JAS07292.1 UFQS01000791 UFQT01000791 SSX06909.1 SSX27253.1 GBHO01023525 GBHO01023524 GDHC01015922 JAG20079.1 JAG20080.1 JAQ02707.1 AJWK01032810 AJWK01032811 JRES01001542 KNC22133.1 ACPB03022158 ACPB03022159 APGK01023024 KB740457 ENN80624.1 CM000363 EDX11204.1 AXCM01005116 CM002912 KMZ00726.1 CH954178 EDV52482.1 M89628 AE014296 BT015959 CM000159 KRK02292.1 CH480826 EDW44181.1 KQS44298.1 KMZ00727.1 KRK02291.1 UFQS01001279 UFQT01001279 SSX10107.1 SSX29829.1 KMZ00725.1 KQS44297.1 EDW95211.1 JXUM01007767 JXUM01007768 JXUM01007769 JXUM01007770 JXUM01007771 JXUM01007772 KQ560247 KXJ83583.1 CH902618 EDV39462.1 KPU78057.1 GAMC01016156 GAMC01016155 JAB90400.1 KPU78056.1 CH963847 EDW72874.2 GAKP01007729 JAC51223.1 OUUW01000009 SPP84721.1 CH379069 KRT07669.1 KRT07673.1 KRT07672.1 SPP84720.1 KRT07671.1 EDY73443.1 CH479188 EDW40193.1 CH940647 EDW70401.1 KRF84894.1 LJIJ01001427 ODM91769.1 CCAG010000237 CP012525 ALC44362.1 CH933809 EDW19411.1 KRG06631.1 CH916366 EDV96600.1 CCAG010004851 KRG06630.1 LNIX01000006 OXA52533.1 IACF01003719 LAB69330.1 GEFH01001259 JAP67322.1 GFPF01011705 MAA22851.1 GEDV01005900 JAP82657.1 GFAA01002374 JAU01061.1 JXJN01007898 JXJN01007899 GFWV01003041 MAA27771.1 GEGO01002314 JAR93090.1
RVE50532.1 AGBW02011268 OWR46920.1 ODYU01002624 SOQ40363.1 NWSH01000662 PCG74944.1 JTDY01003056 KOB70236.1 LJIG01022522 KRT80131.1 KQ971338 EFA01582.2 GECU01030324 JAS77382.1 GEBQ01013744 JAT26233.1 APGK01009376 APGK01009377 KB737932 ENN82832.1 AAAB01008966 EAA12933.4 GFTR01006749 JAW09677.1 CH477268 EAT45428.1 KB632337 ERL92855.1 KK852756 KDR17087.1 DS235760 EEB16582.1 APCN01001959 APCN01001960 ATLV01023923 KE525347 KFB49928.1 GEDC01009827 JAS27471.1 ADMH02000929 ETN64645.1 GEDC01030006 JAS07292.1 UFQS01000791 UFQT01000791 SSX06909.1 SSX27253.1 GBHO01023525 GBHO01023524 GDHC01015922 JAG20079.1 JAG20080.1 JAQ02707.1 AJWK01032810 AJWK01032811 JRES01001542 KNC22133.1 ACPB03022158 ACPB03022159 APGK01023024 KB740457 ENN80624.1 CM000363 EDX11204.1 AXCM01005116 CM002912 KMZ00726.1 CH954178 EDV52482.1 M89628 AE014296 BT015959 CM000159 KRK02292.1 CH480826 EDW44181.1 KQS44298.1 KMZ00727.1 KRK02291.1 UFQS01001279 UFQT01001279 SSX10107.1 SSX29829.1 KMZ00725.1 KQS44297.1 EDW95211.1 JXUM01007767 JXUM01007768 JXUM01007769 JXUM01007770 JXUM01007771 JXUM01007772 KQ560247 KXJ83583.1 CH902618 EDV39462.1 KPU78057.1 GAMC01016156 GAMC01016155 JAB90400.1 KPU78056.1 CH963847 EDW72874.2 GAKP01007729 JAC51223.1 OUUW01000009 SPP84721.1 CH379069 KRT07669.1 KRT07673.1 KRT07672.1 SPP84720.1 KRT07671.1 EDY73443.1 CH479188 EDW40193.1 CH940647 EDW70401.1 KRF84894.1 LJIJ01001427 ODM91769.1 CCAG010000237 CP012525 ALC44362.1 CH933809 EDW19411.1 KRG06631.1 CH916366 EDV96600.1 CCAG010004851 KRG06630.1 LNIX01000006 OXA52533.1 IACF01003719 LAB69330.1 GEFH01001259 JAP67322.1 GFPF01011705 MAA22851.1 GEDV01005900 JAP82657.1 GFAA01002374 JAU01061.1 JXJN01007898 JXJN01007899 GFWV01003041 MAA27771.1 GEGO01002314 JAR93090.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000037510 UP000007266 UP000192223 UP000019118 UP000007062 UP000008820 UP000075881 UP000076407 UP000075920 UP000030742 UP000027135 UP000075900 UP000009046 UP000069272 UP000075884 UP000075840 UP000030765 UP000075885 UP000000673 UP000092461 UP000037069 UP000015103 UP000095301 UP000095300 UP000000304 UP000075903 UP000075883 UP000075880 UP000008711 UP000000803 UP000002282 UP000001292 UP000192221 UP000069940 UP000249989 UP000007801 UP000007798 UP000268350 UP000078200 UP000001819 UP000092445 UP000008744 UP000091820 UP000008792 UP000094527 UP000092444 UP000092553 UP000009192 UP000001070 UP000198287 UP000092460
UP000037510 UP000007266 UP000192223 UP000019118 UP000007062 UP000008820 UP000075881 UP000076407 UP000075920 UP000030742 UP000027135 UP000075900 UP000009046 UP000069272 UP000075884 UP000075840 UP000030765 UP000075885 UP000000673 UP000092461 UP000037069 UP000015103 UP000095301 UP000095300 UP000000304 UP000075903 UP000075883 UP000075880 UP000008711 UP000000803 UP000002282 UP000001292 UP000192221 UP000069940 UP000249989 UP000007801 UP000007798 UP000268350 UP000078200 UP000001819 UP000092445 UP000008744 UP000091820 UP000008792 UP000094527 UP000092444 UP000092553 UP000009192 UP000001070 UP000198287 UP000092460
Pfam
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
H9IUK2
A0A194RLP1
A0A194PJL2
A0A437BJB7
A0A212EZK5
A0A2H1VHP4
+ More
A0A2A4JTJ8 A0A0L7L4I0 A0A0T6AY67 D2A162 A0A1W4WIT1 A0A1B6HRT7 A0A1B6LRB5 N6UVB8 Q7Q2Z9 A0A224XB96 Q17FS3 A0A182K8J4 A0A182WR55 A0A182WF17 A0A1S4F4H5 U4US14 A0A067R243 A0A182RFE2 E0VT76 A0A182FTV4 A0A182NPL6 A0A1S4H6J9 A0A453YJM5 A0A084WI84 A0A182PU84 A0A1B6DP80 W5JP23 A0A1B6C198 A0A336KR11 A0A0A9XJD6 A0A1B0CWS4 A0A0L0BQ75 T1I8U1 A0A1I8ML02 A0A1I8QDX9 N6TJK7 B4QRV8 A0A182V902 A0A182MN75 A0A182IJK1 A0A0J9UPV3 B3NE58 P40421 A0A0R1E3V8 B4IA68 A0A0Q5UKB6 A0A0J9RYJ5 A0A0R1E263 A0A336KZ48 A0A0J9RZW2 A0A0Q5U6J1 B4PFC5 A0A1W4VUR9 A0A182G6D2 B3M4R3 A0A0P8XTZ4 W8ANN1 A0A0P8XTK4 B4MKU9 A0A034WBT5 A0A3B0KHY3 A0A1A9VSD6 A0A0R3P7M8 A0A0R3P3H1 A0A1A9UQS4 A0A0R3P3E4 A0A3B0K9C5 A0A1A9Z5T7 A0A0R3P3C9 B5DPA9 B4GR08 A0A1A9WRH1 B4LG02 A0A0Q9WVD8 A0A1D2MFK2 A0A1B0G8C3 A0A0M4EZY5 A0A1A9Z4V7 B4KV71 A0A0Q9XQA2 B4IYR3 A0A1B0G581 A0A0Q9XDH4 A0A226E4L8 A0A2P2I617 A0A131XLU5 A0A224Z6H9 A0A131YUK5 A0A1E1XP01 A0A1B0B3L8 A0A1A9Z2Z5 A0A293LIW1 A0A147BQQ3
A0A2A4JTJ8 A0A0L7L4I0 A0A0T6AY67 D2A162 A0A1W4WIT1 A0A1B6HRT7 A0A1B6LRB5 N6UVB8 Q7Q2Z9 A0A224XB96 Q17FS3 A0A182K8J4 A0A182WR55 A0A182WF17 A0A1S4F4H5 U4US14 A0A067R243 A0A182RFE2 E0VT76 A0A182FTV4 A0A182NPL6 A0A1S4H6J9 A0A453YJM5 A0A084WI84 A0A182PU84 A0A1B6DP80 W5JP23 A0A1B6C198 A0A336KR11 A0A0A9XJD6 A0A1B0CWS4 A0A0L0BQ75 T1I8U1 A0A1I8ML02 A0A1I8QDX9 N6TJK7 B4QRV8 A0A182V902 A0A182MN75 A0A182IJK1 A0A0J9UPV3 B3NE58 P40421 A0A0R1E3V8 B4IA68 A0A0Q5UKB6 A0A0J9RYJ5 A0A0R1E263 A0A336KZ48 A0A0J9RZW2 A0A0Q5U6J1 B4PFC5 A0A1W4VUR9 A0A182G6D2 B3M4R3 A0A0P8XTZ4 W8ANN1 A0A0P8XTK4 B4MKU9 A0A034WBT5 A0A3B0KHY3 A0A1A9VSD6 A0A0R3P7M8 A0A0R3P3H1 A0A1A9UQS4 A0A0R3P3E4 A0A3B0K9C5 A0A1A9Z5T7 A0A0R3P3C9 B5DPA9 B4GR08 A0A1A9WRH1 B4LG02 A0A0Q9WVD8 A0A1D2MFK2 A0A1B0G8C3 A0A0M4EZY5 A0A1A9Z4V7 B4KV71 A0A0Q9XQA2 B4IYR3 A0A1B0G581 A0A0Q9XDH4 A0A226E4L8 A0A2P2I617 A0A131XLU5 A0A224Z6H9 A0A131YUK5 A0A1E1XP01 A0A1B0B3L8 A0A1A9Z2Z5 A0A293LIW1 A0A147BQQ3
PDB
3H69
E-value=4.56791e-21,
Score=249
Ontologies
GO
Topology
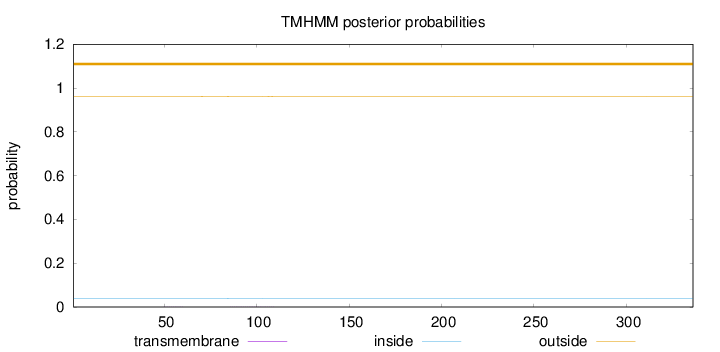
Length:
336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0167
Exp number, first 60 AAs:
0.0024
Total prob of N-in:
0.03933
outside
1 - 336
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
