Gene
KWMTBOMO16738
Pre Gene Modal
BGIBMGA012739
Annotation
PREDICTED:_peroxidase-like?_partial_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.067
Sequence
CDS
ATGGGCTCGAACATATTCCACATATGGTTCTACAGACTCCATAACGTTGTGGCCGCGAATCTGGAGAAGATCAATCCCTGTTGGGATGACAATAAAATTTTCTACACGACGAGAGAAATTCTGATAGCTGGTTATTTACAAATATATTACTATCAATTTTTGCCTCTTTTGTTTGGGATGGAAAGGCTGATAAAAGACGGGGTTATATCAAAACACAAAGGCTACAGAGATGTTTATGATGAGAAGATTATACCACAAATGTCCGACGAGTATTCTTATGTGCTACGATGGTTTCACATTGCACAAGAAGCGACCCTTGAATTGTACGATGAAAACTACAAGTGCTTCAAGACATTCCCTATGATAGCTCGTGGTAGTTTCTTGCAGTCCACTTCTAAATTTATGGACCATGCTGTGGATCCTGATATCTCCGAAGTTGGCTTGGGAGGGTTGCAGTATGCATTCGACATTACAGCTGCGGATGTAGCAAAAGGACGATTATTTGGATTCGCTCCATATGTTGATTATTTGGAAGTATGCAGAAAGAAGAAGTTCAAAACCTTCAAGGATCTTCTTGAAGTGATGGATGAAGATAGAATTAAACGCCTTAGAAGAGTTTACAAATGTGCACAAGACGTAGACCTGATGCCTGGGCTATGGTCTGAGCGAAAAATAAAGGATGGAAGTGTCCCGGTACCCTATACTGCCTAA
Protein
MGSNIFHIWFYRLHNVVAANLEKINPCWDDNKIFYTTREILIAGYLQIYYYQFLPLLFGMERLIKDGVISKHKGYRDVYDEKIIPQMSDEYSYVLRWFHIAQEATLELYDENYKCFKTFPMIARGSFLQSTSKFMDHAVDPDISEVGLGGLQYAFDITAADVAKGRLFGFAPYVDYLEVCRKKKFKTFKDLLEVMDEDRIKRLRRVYKCAQDVDLMPGLWSERKIKDGSVPVPYTA
Summary
Uniprot
H9JT77
I4DS54
A0A194RBC8
A0A194Q7Y0
A0A194QDV6
A0A194Q9H2
+ More
I4DJ10 A0A3S2NLC3 A0A194RAP6 A0A2H1WGR9 A0A2W1BYE5 A0A2A4IUC0 A0A212ETD1 A0A2H1WBG2 H9ISL1 A0A2W1BT67 A0A2H1W4U4 A0A2H1W8U7 A0A2H1W9U4 A0A2W1BT77 H9JT78 A0A2A4JI21 A0A088MGW5 A0A2A4J7D3 H9IT78 A0A2H1VUX0 H9JQT0 A0A194Q3N0 A0A437AS20 A0A437ASV3 A0A437AYB0 A0A3S2NMV9 A0A3S2L850 A0A0N0PED8 H9ISL2 A0A2H1UZX5 A0A2W1BXH1 A0A2W1BQ93 A0A2A4IUP3 A0A2W1BS42 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 A0A3S2M0V1 A0A0P4VM15 T1I7W3 A0A0L7LKV1 A0A1I7XWL8 A0A1I7XW56 A0A0P5K1Q1 A0A2P2IA20 A0A0P5IMW3 A0A2H8TD85 A0A0P6F9A4 A0A0P6E141 A0A0N8DZX3 A0A1D2NCP0 A0A0N8A5Q2 A0A0P5HUB2 A0A0M3KBE9 A0A3P6RD52 A0A2A2JDR5 A0A1B6DIG5 A0A0P5XNL2 A0A0M3IZG9 A0A0P5BFF7 A0A0N8BD57 A0A0P5LIN7 A0A0P5XJJ1 A0A0P5TGT2 A0A0P5TXW4 A0A0P5AGR2 A0A1B6CUT2 A0A0N8CQ46 A0A0P5MWS3 A0A0P5XAG5 A0A0P5TQQ2 A0A0P5MAF8 A0A0P5AGT5 A0A0P5W887 A0A0P5Q006 A0A0P5L589 A0A0P5ZJN5 A0A0P5NBQ6 A0A0P5JUL6 A0A2A2L2J1 A0A0P6DEV7 A0A0N8C7Q8 A0A0P5TWY8 A0A0N7ZYS0 A0A0P6EXI3 A0A0P6CK58 A0A0P5ERW6 A0A0P6I0W9 A0A0A9ZAS3 G3MHL8 A0A146M0F8
I4DJ10 A0A3S2NLC3 A0A194RAP6 A0A2H1WGR9 A0A2W1BYE5 A0A2A4IUC0 A0A212ETD1 A0A2H1WBG2 H9ISL1 A0A2W1BT67 A0A2H1W4U4 A0A2H1W8U7 A0A2H1W9U4 A0A2W1BT77 H9JT78 A0A2A4JI21 A0A088MGW5 A0A2A4J7D3 H9IT78 A0A2H1VUX0 H9JQT0 A0A194Q3N0 A0A437AS20 A0A437ASV3 A0A437AYB0 A0A3S2NMV9 A0A3S2L850 A0A0N0PED8 H9ISL2 A0A2H1UZX5 A0A2W1BXH1 A0A2W1BQ93 A0A2A4IUP3 A0A2W1BS42 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 A0A3S2M0V1 A0A0P4VM15 T1I7W3 A0A0L7LKV1 A0A1I7XWL8 A0A1I7XW56 A0A0P5K1Q1 A0A2P2IA20 A0A0P5IMW3 A0A2H8TD85 A0A0P6F9A4 A0A0P6E141 A0A0N8DZX3 A0A1D2NCP0 A0A0N8A5Q2 A0A0P5HUB2 A0A0M3KBE9 A0A3P6RD52 A0A2A2JDR5 A0A1B6DIG5 A0A0P5XNL2 A0A0M3IZG9 A0A0P5BFF7 A0A0N8BD57 A0A0P5LIN7 A0A0P5XJJ1 A0A0P5TGT2 A0A0P5TXW4 A0A0P5AGR2 A0A1B6CUT2 A0A0N8CQ46 A0A0P5MWS3 A0A0P5XAG5 A0A0P5TQQ2 A0A0P5MAF8 A0A0P5AGT5 A0A0P5W887 A0A0P5Q006 A0A0P5L589 A0A0P5ZJN5 A0A0P5NBQ6 A0A0P5JUL6 A0A2A2L2J1 A0A0P6DEV7 A0A0N8C7Q8 A0A0P5TWY8 A0A0N7ZYS0 A0A0P6EXI3 A0A0P6CK58 A0A0P5ERW6 A0A0P6I0W9 A0A0A9ZAS3 G3MHL8 A0A146M0F8
Pubmed
EMBL
BABH01034743
BABH01034744
AK405426
BAM20744.1
KQ460416
KPJ14922.1
+ More
KQ459324 KPJ01647.1 KPJ01646.1 KPJ01645.1 AK401278 BAM17900.1 RSAL01000046 RVE50539.1 KPJ14923.1 ODYU01008556 SOQ52260.1 KZ149907 PZC78257.1 NWSH01006580 PCG63355.1 AGBW02012596 OWR44740.1 ODYU01007479 SOQ50286.1 BABH01013455 PZC78258.1 ODYU01006337 SOQ48101.1 ODYU01007063 SOQ49501.1 ODYU01007226 SOQ49823.1 PZC78259.1 BABH01034746 BABH01034747 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 NWSH01002854 PCG67434.1 BABH01013458 BABH01013459 BABH01013460 ODYU01004596 SOQ44641.1 BABH01028746 BABH01028747 BABH01028748 KQ459564 KPI99614.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 RSAL01000273 RVE43156.1 RSAL01000231 RVE43837.1 RVE41111.1 KQ459837 KPJ19761.1 BABH01013447 BABH01013448 ODYU01000046 SOQ34155.1 KZ149956 PZC76463.1 PZC76461.1 NWSH01006414 PCG63451.1 PZC76464.1 NWSH01006200 PCG63597.1 NWSH01001116 PCG72533.1 PCG72532.1 RSAL01000083 RVE48485.1 GDKW01003332 JAI53263.1 ACPB03011632 JTDY01000732 KOB76072.1 GDIQ01192196 JAK59529.1 IACF01005278 LAB70864.1 GDIQ01210487 JAK41238.1 GFXV01000266 MBW12071.1 GDIQ01053609 JAN41128.1 GDIQ01070734 JAN24003.1 GDIQ01075484 JAN19253.1 LJIJ01000089 ODN03012.1 GDIP01179921 JAJ43481.1 GDIQ01227980 JAK23745.1 UYRR01034464 VDK61142.1 LIAE01010510 PAV59662.1 GEDC01011809 JAS25489.1 GDIP01069488 JAM34227.1 UYRR01000388 VDK17833.1 GDIP01185931 JAJ37471.1 GDIQ01189403 JAK62322.1 GDIQ01173053 JAK78672.1 GDIP01071190 JAM32525.1 GDIQ01091829 JAL59897.1 GDIP01122932 JAL80782.1 GDIP01199332 JAJ24070.1 GEDC01020121 JAS17177.1 GDIP01109879 JAL93835.1 GDIQ01150074 JAL01652.1 GDIP01074949 JAM28766.1 GDIP01122937 JAL80777.1 GDIQ01161021 JAK90704.1 GDIP01199331 JAJ24071.1 GDIP01090990 JAM12725.1 GDIQ01123279 JAL28447.1 GDIQ01175401 JAK76324.1 GDIP01044106 JAM59609.1 GDIQ01155217 JAK96508.1 GDIQ01219353 GDIQ01064750 GDIQ01030254 JAK32372.1 LIAE01007263 PAV80481.1 GDIQ01079931 JAN14806.1 GDIQ01106595 JAL45131.1 GDIP01122933 JAL80781.1 GDIP01199377 JAJ24025.1 GDIQ01069105 JAN25632.1 GDIP01007020 JAM96695.1 GDIQ01268388 JAJ83336.1 GDIQ01012997 JAN81740.1 GBHO01004699 JAG38905.1 JO841369 AEO32986.1 GDHC01005325 JAQ13304.1
KQ459324 KPJ01647.1 KPJ01646.1 KPJ01645.1 AK401278 BAM17900.1 RSAL01000046 RVE50539.1 KPJ14923.1 ODYU01008556 SOQ52260.1 KZ149907 PZC78257.1 NWSH01006580 PCG63355.1 AGBW02012596 OWR44740.1 ODYU01007479 SOQ50286.1 BABH01013455 PZC78258.1 ODYU01006337 SOQ48101.1 ODYU01007063 SOQ49501.1 ODYU01007226 SOQ49823.1 PZC78259.1 BABH01034746 BABH01034747 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 NWSH01002854 PCG67434.1 BABH01013458 BABH01013459 BABH01013460 ODYU01004596 SOQ44641.1 BABH01028746 BABH01028747 BABH01028748 KQ459564 KPI99614.1 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 RSAL01000273 RVE43156.1 RSAL01000231 RVE43837.1 RVE41111.1 KQ459837 KPJ19761.1 BABH01013447 BABH01013448 ODYU01000046 SOQ34155.1 KZ149956 PZC76463.1 PZC76461.1 NWSH01006414 PCG63451.1 PZC76464.1 NWSH01006200 PCG63597.1 NWSH01001116 PCG72533.1 PCG72532.1 RSAL01000083 RVE48485.1 GDKW01003332 JAI53263.1 ACPB03011632 JTDY01000732 KOB76072.1 GDIQ01192196 JAK59529.1 IACF01005278 LAB70864.1 GDIQ01210487 JAK41238.1 GFXV01000266 MBW12071.1 GDIQ01053609 JAN41128.1 GDIQ01070734 JAN24003.1 GDIQ01075484 JAN19253.1 LJIJ01000089 ODN03012.1 GDIP01179921 JAJ43481.1 GDIQ01227980 JAK23745.1 UYRR01034464 VDK61142.1 LIAE01010510 PAV59662.1 GEDC01011809 JAS25489.1 GDIP01069488 JAM34227.1 UYRR01000388 VDK17833.1 GDIP01185931 JAJ37471.1 GDIQ01189403 JAK62322.1 GDIQ01173053 JAK78672.1 GDIP01071190 JAM32525.1 GDIQ01091829 JAL59897.1 GDIP01122932 JAL80782.1 GDIP01199332 JAJ24070.1 GEDC01020121 JAS17177.1 GDIP01109879 JAL93835.1 GDIQ01150074 JAL01652.1 GDIP01074949 JAM28766.1 GDIP01122937 JAL80777.1 GDIQ01161021 JAK90704.1 GDIP01199331 JAJ24071.1 GDIP01090990 JAM12725.1 GDIQ01123279 JAL28447.1 GDIQ01175401 JAK76324.1 GDIP01044106 JAM59609.1 GDIQ01155217 JAK96508.1 GDIQ01219353 GDIQ01064750 GDIQ01030254 JAK32372.1 LIAE01007263 PAV80481.1 GDIQ01079931 JAN14806.1 GDIQ01106595 JAL45131.1 GDIP01122933 JAL80781.1 GDIP01199377 JAJ24025.1 GDIQ01069105 JAN25632.1 GDIP01007020 JAM96695.1 GDIQ01268388 JAJ83336.1 GDIQ01012997 JAN81740.1 GBHO01004699 JAG38905.1 JO841369 AEO32986.1 GDHC01005325 JAQ13304.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JT77
I4DS54
A0A194RBC8
A0A194Q7Y0
A0A194QDV6
A0A194Q9H2
+ More
I4DJ10 A0A3S2NLC3 A0A194RAP6 A0A2H1WGR9 A0A2W1BYE5 A0A2A4IUC0 A0A212ETD1 A0A2H1WBG2 H9ISL1 A0A2W1BT67 A0A2H1W4U4 A0A2H1W8U7 A0A2H1W9U4 A0A2W1BT77 H9JT78 A0A2A4JI21 A0A088MGW5 A0A2A4J7D3 H9IT78 A0A2H1VUX0 H9JQT0 A0A194Q3N0 A0A437AS20 A0A437ASV3 A0A437AYB0 A0A3S2NMV9 A0A3S2L850 A0A0N0PED8 H9ISL2 A0A2H1UZX5 A0A2W1BXH1 A0A2W1BQ93 A0A2A4IUP3 A0A2W1BS42 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 A0A3S2M0V1 A0A0P4VM15 T1I7W3 A0A0L7LKV1 A0A1I7XWL8 A0A1I7XW56 A0A0P5K1Q1 A0A2P2IA20 A0A0P5IMW3 A0A2H8TD85 A0A0P6F9A4 A0A0P6E141 A0A0N8DZX3 A0A1D2NCP0 A0A0N8A5Q2 A0A0P5HUB2 A0A0M3KBE9 A0A3P6RD52 A0A2A2JDR5 A0A1B6DIG5 A0A0P5XNL2 A0A0M3IZG9 A0A0P5BFF7 A0A0N8BD57 A0A0P5LIN7 A0A0P5XJJ1 A0A0P5TGT2 A0A0P5TXW4 A0A0P5AGR2 A0A1B6CUT2 A0A0N8CQ46 A0A0P5MWS3 A0A0P5XAG5 A0A0P5TQQ2 A0A0P5MAF8 A0A0P5AGT5 A0A0P5W887 A0A0P5Q006 A0A0P5L589 A0A0P5ZJN5 A0A0P5NBQ6 A0A0P5JUL6 A0A2A2L2J1 A0A0P6DEV7 A0A0N8C7Q8 A0A0P5TWY8 A0A0N7ZYS0 A0A0P6EXI3 A0A0P6CK58 A0A0P5ERW6 A0A0P6I0W9 A0A0A9ZAS3 G3MHL8 A0A146M0F8
I4DJ10 A0A3S2NLC3 A0A194RAP6 A0A2H1WGR9 A0A2W1BYE5 A0A2A4IUC0 A0A212ETD1 A0A2H1WBG2 H9ISL1 A0A2W1BT67 A0A2H1W4U4 A0A2H1W8U7 A0A2H1W9U4 A0A2W1BT77 H9JT78 A0A2A4JI21 A0A088MGW5 A0A2A4J7D3 H9IT78 A0A2H1VUX0 H9JQT0 A0A194Q3N0 A0A437AS20 A0A437ASV3 A0A437AYB0 A0A3S2NMV9 A0A3S2L850 A0A0N0PED8 H9ISL2 A0A2H1UZX5 A0A2W1BXH1 A0A2W1BQ93 A0A2A4IUP3 A0A2W1BS42 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 A0A3S2M0V1 A0A0P4VM15 T1I7W3 A0A0L7LKV1 A0A1I7XWL8 A0A1I7XW56 A0A0P5K1Q1 A0A2P2IA20 A0A0P5IMW3 A0A2H8TD85 A0A0P6F9A4 A0A0P6E141 A0A0N8DZX3 A0A1D2NCP0 A0A0N8A5Q2 A0A0P5HUB2 A0A0M3KBE9 A0A3P6RD52 A0A2A2JDR5 A0A1B6DIG5 A0A0P5XNL2 A0A0M3IZG9 A0A0P5BFF7 A0A0N8BD57 A0A0P5LIN7 A0A0P5XJJ1 A0A0P5TGT2 A0A0P5TXW4 A0A0P5AGR2 A0A1B6CUT2 A0A0N8CQ46 A0A0P5MWS3 A0A0P5XAG5 A0A0P5TQQ2 A0A0P5MAF8 A0A0P5AGT5 A0A0P5W887 A0A0P5Q006 A0A0P5L589 A0A0P5ZJN5 A0A0P5NBQ6 A0A0P5JUL6 A0A2A2L2J1 A0A0P6DEV7 A0A0N8C7Q8 A0A0P5TWY8 A0A0N7ZYS0 A0A0P6EXI3 A0A0P6CK58 A0A0P5ERW6 A0A0P6I0W9 A0A0A9ZAS3 G3MHL8 A0A146M0F8
PDB
3R5Q
E-value=8.51786e-14,
Score=184
Ontologies
GO
PANTHER
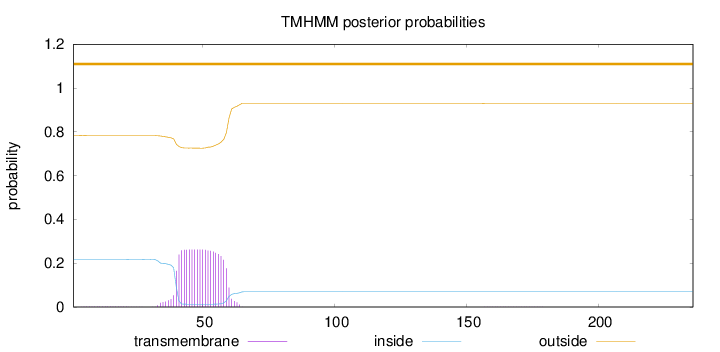
Topology
Length:
236
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.31625999999999
Exp number, first 60 AAs:
5.21041
Total prob of N-in:
0.21595
outside
1 - 236
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
