Gene
KWMTBOMO16737 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012740
Annotation
PREDICTED:_peroxidase-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.667 PlasmaMembrane Reliability : 1.624
Sequence
CDS
ATGCTCTGGTCATTAGTTTTCTTGGCGTTTCTGAAGCTGACCGCCGGTGTTCGCTATGAATCGTTTTCTGGTCTACCAGCGACTGATGAAAAGATCCAATACTTCATCAAACAGGAAAATTTAGATAACTGCACCAATGACATTCAACTCCCCTGCGACCCCAATGAGAGACGACGTCTAGATGGCTCATGCAGTAATTTCGACTATCCGTCAAGAGGAACATTCCATACCCCTGTGATAAGATTGTTACCCGAAGCCGACTGTGGAGACGATGAAACCTCTCCATCTGGAGCTCCTTTAAAATCTGCCCGGGAAGTCAGGCAGCGTATTTTACAGACAGGGAAGGCATCAGACCTCTCATACACACAACTATTGGCGATTATCTCGACAATCATATTTGCTGACCTTGGTTCAATCCATGATTCCGTGAATTTGTTAACGGAGACCACAAATTGTTGCACAGCGGAAGGAAAATCAAATTATATGTGCACGCCTATAGATATTCCCCAGGACGACCCTGTCCATAGGTTCTCTGGTATACGCTGCTTGAATCTCACTCGACCGAAATCGTTCCAGACTTATGGCTGCCTTGCTGACTCGAATGTAGACAGAATCGAATTCACAACACCGTTATTTGATTTATCGACTATCTACCGATCGACCGAACCGGCTCCAGAATACAGAACATACAGTGGCGGATTACTGATGACCCAAGAGGCCGATGGCACCATCTTCCCCCCACAAGAAGGACCGCACAGCAATAAATGCTTGCAAAACGACGCGTCTAATGGAGAAACGAAATGTTTTGGCCCTGTTTCAACATCGATTCTGCCGGTCACATTGTTGGTCGTGTGGTGGTGGAGACTACACAATAAGATCGCCAAAGAACTAACGAAATCAATCCTCATTGGGATGACGAAACATTGTTCCAAACTGCAAGGGACATTAACATCGCAATAA
Protein
MLWSLVFLAFLKLTAGVRYESFSGLPATDEKIQYFIKQENLDNCTNDIQLPCDPNERRRLDGSCSNFDYPSRGTFHTPVIRLLPEADCGDDETSPSGAPLKSAREVRQRILQTGKASDLSYTQLLAIISTIIFADLGSIHDSVNLLTETTNCCTAEGKSNYMCTPIDIPQDDPVHRFSGIRCLNLTRPKSFQTYGCLADSNVDRIEFTTPLFDLSTIYRSTEPAPEYRTYSGGLLMTQEADGTIFPPQEGPHSNKCLQNDASNGETKCFGPVSTSILPVTLLVVWWWRLHNKIAKELTKSILIGMTKHCSKLQGTLTSQ
Summary
Uniprot
H9JT78
A0A2H1WBG2
A0A2H1WAX3
A0A2W1BT67
A0A3S2NLC3
A0A2H1W9U4
+ More
A0A2A4J7D3 H9JT77 A0A2A4IUC0 I4DJ10 A0A2H1W2N9 A0A194QDV6 A0A212ETD1 A0A194RAP6 A0A2H1VUX0 A0A2W1BYE5 A0A194Q7Y0 A0A194Q9H2 H9JQT0 A0A2H1W8U7 A0A2W1BT77 A0A194R5X9 A0A088MGW5 A0A437AU04 A0A194Q3N0 A0A437BBT0 A0A2W1BQ93 A0A2A4JI21 A0A2A4JMB0 A0A2W1BS42 A0A437AYB0 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 A0A2H1UZX5 A0A2M4ABL8 A0A2M4AAY9 W5JRW3 A0A1Y9G8K4 A0A2J7RAM0 A0A2M4DQF3 A0A2M3ZYF0 A0A2M4BI03 A0A1Y9G8H0 W5JTG7 B6DDU6 Q9XYP9 A0A3S2M8S7 A0A2H1W2P7 H9JAH5 A0A2M3ZY83 A0A2M3ZXR4 A0A2M3ZXX0 A0A067RJX3 T1IHP0 A0A346D9R6 A0A2M3ZXZ6 A0A2P8XQD7 A0A088MG66 A0A2A4JJ63 A0A1S3D3U1 A0A088SIJ3 A0A0C9R1B3 A0A140GY71 A0A0P5XSE0 A0A0P5F515 A0A0P6A0Z4 A0A0P5URR0 A0A0P5UPH1 A0A0P5E8L4 A0A0P5Z353 A0A0P5A5P4 A0A0P5TSR8 A0A0P6HK49 A0A0P5DST8 A0A0P5IXM5 A0A0P5SC50 A0A0N8AGZ6 A0A0P5TTP5 A0A0P4Z5Z3 A0A0P4WNF7 A0A0P5T2Q4 A0A0P5ZJM0 A0A0P4YC91
A0A2A4J7D3 H9JT77 A0A2A4IUC0 I4DJ10 A0A2H1W2N9 A0A194QDV6 A0A212ETD1 A0A194RAP6 A0A2H1VUX0 A0A2W1BYE5 A0A194Q7Y0 A0A194Q9H2 H9JQT0 A0A2H1W8U7 A0A2W1BT77 A0A194R5X9 A0A088MGW5 A0A437AU04 A0A194Q3N0 A0A437BBT0 A0A2W1BQ93 A0A2A4JI21 A0A2A4JMB0 A0A2W1BS42 A0A437AYB0 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 A0A2H1UZX5 A0A2M4ABL8 A0A2M4AAY9 W5JRW3 A0A1Y9G8K4 A0A2J7RAM0 A0A2M4DQF3 A0A2M3ZYF0 A0A2M4BI03 A0A1Y9G8H0 W5JTG7 B6DDU6 Q9XYP9 A0A3S2M8S7 A0A2H1W2P7 H9JAH5 A0A2M3ZY83 A0A2M3ZXR4 A0A2M3ZXX0 A0A067RJX3 T1IHP0 A0A346D9R6 A0A2M3ZXZ6 A0A2P8XQD7 A0A088MG66 A0A2A4JJ63 A0A1S3D3U1 A0A088SIJ3 A0A0C9R1B3 A0A140GY71 A0A0P5XSE0 A0A0P5F515 A0A0P6A0Z4 A0A0P5URR0 A0A0P5UPH1 A0A0P5E8L4 A0A0P5Z353 A0A0P5A5P4 A0A0P5TSR8 A0A0P6HK49 A0A0P5DST8 A0A0P5IXM5 A0A0P5SC50 A0A0N8AGZ6 A0A0P5TTP5 A0A0P4Z5Z3 A0A0P4WNF7 A0A0P5T2Q4 A0A0P5ZJM0 A0A0P4YC91
Pubmed
EMBL
BABH01034746
BABH01034747
ODYU01007479
SOQ50286.1
ODYU01007325
SOQ50002.1
+ More
KZ149907 PZC78258.1 RSAL01000046 RVE50539.1 ODYU01007226 SOQ49823.1 NWSH01002854 PCG67434.1 BABH01034743 BABH01034744 NWSH01006580 PCG63355.1 AK401278 BAM17900.1 ODYU01005947 SOQ47370.1 KQ459324 KPJ01646.1 AGBW02012596 OWR44740.1 KQ460416 KPJ14923.1 ODYU01004596 SOQ44641.1 PZC78257.1 KPJ01647.1 KPJ01645.1 BABH01028746 BABH01028747 BABH01028748 ODYU01007063 SOQ49501.1 PZC78259.1 KQ460685 KPJ12919.1 KJ995811 AIN39494.1 RSAL01000538 RVE41417.1 KQ459564 KPI99614.1 RSAL01000094 RVE47835.1 KZ149956 PZC76461.1 NWSH01001320 PCG71707.1 NWSH01001116 PCG72532.1 PZC76464.1 RSAL01000273 RVE43156.1 PCG72533.1 NWSH01006414 PCG63451.1 PZC76463.1 NWSH01006200 PCG63597.1 ODYU01000046 SOQ34155.1 GGFK01004863 MBW38184.1 GGFK01004599 MBW37920.1 ADMH02000540 ETN66033.1 NEVH01006567 PNF37876.1 GGFL01015608 MBW79786.1 GGFK01000169 MBW33490.1 GGFJ01003549 MBW52690.1 ETN66034.1 EU934306 ACI30084.1 AF118391 AAD22196.1 RSAL01000011 RVE53586.1 ODYU01005682 SOQ46784.1 BABH01009341 GGFK01000027 MBW33348.1 GGFK01000023 MBW33344.1 GGFK01000024 MBW33345.1 KK852427 KDR24107.1 JH429957 MH606240 AXM43807.1 GGFK01000125 MBW33446.1 PYGN01001533 PSN34226.1 KJ995807 AIN39490.1 NWSH01001238 PCG72011.1 KM384736 AIO11227.1 GBYB01000656 JAG70423.1 KT825861 AMN85760.1 GDIP01069925 JAM33790.1 GDIQ01267596 JAJ84128.1 GDIP01040529 JAM63186.1 GDIP01109325 JAL94389.1 GDIP01127616 JAL76098.1 GDIP01163415 JAJ59987.1 GDIP01050634 JAM53081.1 GDIP01203711 JAJ19691.1 GDIP01253153 GDIP01251953 GDIP01201971 GDIP01176159 GDIP01160066 GDIP01140068 GDIP01095332 GDIP01094076 GDIP01074225 JAL63646.1 GDIQ01041401 JAN53336.1 GDIP01253503 GDIP01220810 GDIP01184165 GDIP01167363 GDIP01166614 GDIP01156067 GDIP01153368 GDIP01137919 GDIP01092221 GDIP01088322 GDIP01065029 JAJ70034.1 GDIQ01208932 JAK42793.1 GDIQ01097974 JAL53752.1 GDIP01237962 GDIP01232011 GDIP01221860 GDIP01199311 GDIP01148369 GDIP01097628 GDIP01084673 GDIP01072781 GDIP01062579 JAJ75033.1 GDIP01125618 GDIP01122117 JAL81597.1 GDIP01218159 JAJ05243.1 GDIP01253003 GDIP01134420 GDIP01133121 GDIP01045678 JAI70398.1 GDIP01240365 GDIP01131870 JAL71844.1 GDIP01239257 GDIP01205868 GDIP01149718 GDIP01141176 GDIP01044133 JAM59582.1 GDIP01233156 JAI90245.1
KZ149907 PZC78258.1 RSAL01000046 RVE50539.1 ODYU01007226 SOQ49823.1 NWSH01002854 PCG67434.1 BABH01034743 BABH01034744 NWSH01006580 PCG63355.1 AK401278 BAM17900.1 ODYU01005947 SOQ47370.1 KQ459324 KPJ01646.1 AGBW02012596 OWR44740.1 KQ460416 KPJ14923.1 ODYU01004596 SOQ44641.1 PZC78257.1 KPJ01647.1 KPJ01645.1 BABH01028746 BABH01028747 BABH01028748 ODYU01007063 SOQ49501.1 PZC78259.1 KQ460685 KPJ12919.1 KJ995811 AIN39494.1 RSAL01000538 RVE41417.1 KQ459564 KPI99614.1 RSAL01000094 RVE47835.1 KZ149956 PZC76461.1 NWSH01001320 PCG71707.1 NWSH01001116 PCG72532.1 PZC76464.1 RSAL01000273 RVE43156.1 PCG72533.1 NWSH01006414 PCG63451.1 PZC76463.1 NWSH01006200 PCG63597.1 ODYU01000046 SOQ34155.1 GGFK01004863 MBW38184.1 GGFK01004599 MBW37920.1 ADMH02000540 ETN66033.1 NEVH01006567 PNF37876.1 GGFL01015608 MBW79786.1 GGFK01000169 MBW33490.1 GGFJ01003549 MBW52690.1 ETN66034.1 EU934306 ACI30084.1 AF118391 AAD22196.1 RSAL01000011 RVE53586.1 ODYU01005682 SOQ46784.1 BABH01009341 GGFK01000027 MBW33348.1 GGFK01000023 MBW33344.1 GGFK01000024 MBW33345.1 KK852427 KDR24107.1 JH429957 MH606240 AXM43807.1 GGFK01000125 MBW33446.1 PYGN01001533 PSN34226.1 KJ995807 AIN39490.1 NWSH01001238 PCG72011.1 KM384736 AIO11227.1 GBYB01000656 JAG70423.1 KT825861 AMN85760.1 GDIP01069925 JAM33790.1 GDIQ01267596 JAJ84128.1 GDIP01040529 JAM63186.1 GDIP01109325 JAL94389.1 GDIP01127616 JAL76098.1 GDIP01163415 JAJ59987.1 GDIP01050634 JAM53081.1 GDIP01203711 JAJ19691.1 GDIP01253153 GDIP01251953 GDIP01201971 GDIP01176159 GDIP01160066 GDIP01140068 GDIP01095332 GDIP01094076 GDIP01074225 JAL63646.1 GDIQ01041401 JAN53336.1 GDIP01253503 GDIP01220810 GDIP01184165 GDIP01167363 GDIP01166614 GDIP01156067 GDIP01153368 GDIP01137919 GDIP01092221 GDIP01088322 GDIP01065029 JAJ70034.1 GDIQ01208932 JAK42793.1 GDIQ01097974 JAL53752.1 GDIP01237962 GDIP01232011 GDIP01221860 GDIP01199311 GDIP01148369 GDIP01097628 GDIP01084673 GDIP01072781 GDIP01062579 JAJ75033.1 GDIP01125618 GDIP01122117 JAL81597.1 GDIP01218159 JAJ05243.1 GDIP01253003 GDIP01134420 GDIP01133121 GDIP01045678 JAI70398.1 GDIP01240365 GDIP01131870 JAL71844.1 GDIP01239257 GDIP01205868 GDIP01149718 GDIP01141176 GDIP01044133 JAM59582.1 GDIP01233156 JAI90245.1
Proteomes
Pfam
PF03098 An_peroxidase
Interpro
SUPFAM
SSF48113
SSF48113
Gene 3D
ProteinModelPortal
H9JT78
A0A2H1WBG2
A0A2H1WAX3
A0A2W1BT67
A0A3S2NLC3
A0A2H1W9U4
+ More
A0A2A4J7D3 H9JT77 A0A2A4IUC0 I4DJ10 A0A2H1W2N9 A0A194QDV6 A0A212ETD1 A0A194RAP6 A0A2H1VUX0 A0A2W1BYE5 A0A194Q7Y0 A0A194Q9H2 H9JQT0 A0A2H1W8U7 A0A2W1BT77 A0A194R5X9 A0A088MGW5 A0A437AU04 A0A194Q3N0 A0A437BBT0 A0A2W1BQ93 A0A2A4JI21 A0A2A4JMB0 A0A2W1BS42 A0A437AYB0 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 A0A2H1UZX5 A0A2M4ABL8 A0A2M4AAY9 W5JRW3 A0A1Y9G8K4 A0A2J7RAM0 A0A2M4DQF3 A0A2M3ZYF0 A0A2M4BI03 A0A1Y9G8H0 W5JTG7 B6DDU6 Q9XYP9 A0A3S2M8S7 A0A2H1W2P7 H9JAH5 A0A2M3ZY83 A0A2M3ZXR4 A0A2M3ZXX0 A0A067RJX3 T1IHP0 A0A346D9R6 A0A2M3ZXZ6 A0A2P8XQD7 A0A088MG66 A0A2A4JJ63 A0A1S3D3U1 A0A088SIJ3 A0A0C9R1B3 A0A140GY71 A0A0P5XSE0 A0A0P5F515 A0A0P6A0Z4 A0A0P5URR0 A0A0P5UPH1 A0A0P5E8L4 A0A0P5Z353 A0A0P5A5P4 A0A0P5TSR8 A0A0P6HK49 A0A0P5DST8 A0A0P5IXM5 A0A0P5SC50 A0A0N8AGZ6 A0A0P5TTP5 A0A0P4Z5Z3 A0A0P4WNF7 A0A0P5T2Q4 A0A0P5ZJM0 A0A0P4YC91
A0A2A4J7D3 H9JT77 A0A2A4IUC0 I4DJ10 A0A2H1W2N9 A0A194QDV6 A0A212ETD1 A0A194RAP6 A0A2H1VUX0 A0A2W1BYE5 A0A194Q7Y0 A0A194Q9H2 H9JQT0 A0A2H1W8U7 A0A2W1BT77 A0A194R5X9 A0A088MGW5 A0A437AU04 A0A194Q3N0 A0A437BBT0 A0A2W1BQ93 A0A2A4JI21 A0A2A4JMB0 A0A2W1BS42 A0A437AYB0 A0A2A4JM77 A0A2A4IUP3 A0A2W1BXH1 A0A2A4IUD5 A0A2H1UZX5 A0A2M4ABL8 A0A2M4AAY9 W5JRW3 A0A1Y9G8K4 A0A2J7RAM0 A0A2M4DQF3 A0A2M3ZYF0 A0A2M4BI03 A0A1Y9G8H0 W5JTG7 B6DDU6 Q9XYP9 A0A3S2M8S7 A0A2H1W2P7 H9JAH5 A0A2M3ZY83 A0A2M3ZXR4 A0A2M3ZXX0 A0A067RJX3 T1IHP0 A0A346D9R6 A0A2M3ZXZ6 A0A2P8XQD7 A0A088MG66 A0A2A4JJ63 A0A1S3D3U1 A0A088SIJ3 A0A0C9R1B3 A0A140GY71 A0A0P5XSE0 A0A0P5F515 A0A0P6A0Z4 A0A0P5URR0 A0A0P5UPH1 A0A0P5E8L4 A0A0P5Z353 A0A0P5A5P4 A0A0P5TSR8 A0A0P6HK49 A0A0P5DST8 A0A0P5IXM5 A0A0P5SC50 A0A0N8AGZ6 A0A0P5TTP5 A0A0P4Z5Z3 A0A0P4WNF7 A0A0P5T2Q4 A0A0P5ZJM0 A0A0P4YC91
Ontologies
GO
PANTHER
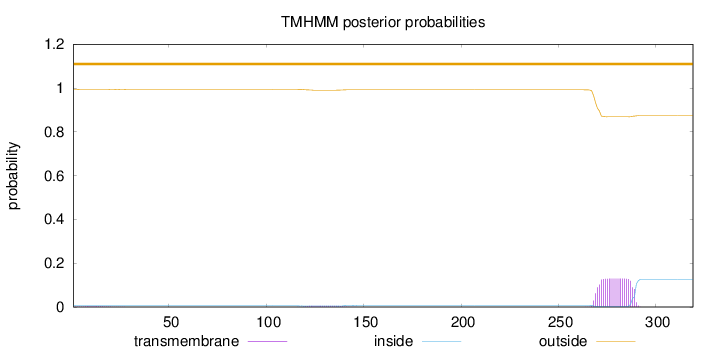
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.982412

Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.70204
Exp number, first 60 AAs:
0.04631
Total prob of N-in:
0.00739
outside
1 - 319
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
