Gene
KWMTBOMO16731
Annotation
PREDICTED:_uncharacterized_protein_LOC106105382_isoform_X2_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.72
Sequence
CDS
ATGAGAGGCTTGAGGTTAGCCACCAGAGACACTGATGCTTTTACGGATCATGCCTTTGAGGTTATCAGAGATGAAATAAACTCAAGCTTACTCCAACCGGAGCTCAGTGATGTTGTGGTCAAGATACCGGGGTCGGTGTCCGTTTCCAATAGGAGGGTCAGCTTCGTGGTGTTCAGAACTGACCGCGCCTTCAGAAATCCAGGGTCCGAGTATTCCGTGAACAGCAGAGTCGTCAGCATCAAGATAGGGAACCTGACGCGGTTTCAAAACGAGGAGTATGATTGTAATTTTACTGATGATGTTAATGTTAAAAGCAAGAGTGTCATGATCCTGGGAGACTCCATCGGCTGCAGTGCTATCAAACGTCCAACAAATGACTGTCGTTTGTTCCAGTTTATCGACATTTATCTGAATCCAATAGTGGAACTGCGCGGGAGCAAGCAAAAGCGAGTTTGCGCGTACTGGGAGTTCCTAGAGGACGGAACAGGGTACTGGTCACAGGACGGATGTGTTCTGATGCGCTCTGAAGATGAAGGGATGCTGGATTACTGCCGATGCACTCATCTGACACACTTCGCTGAGATTCTCATACCTAGGACGTCCTTCTCTGACGCCCACGAAAACGCTCTCGAGCTGATCTCTGTGATCGGCTGCTGTGTCTCCATTTTAGGGCTGGTCTTGGTCGCCATCACCGCGATCCTCTTCAGGTCCTGGCGCCGGGATTTCACCAACAAAATCTGGCTGCAGCTCTGCCTCGCTATATTCATAACCAGCGTCTGCTTCCTCGTCATCGTGTTCGCTGACTTCCTGGAGTACAACATCGCTTGCATGCTGGTCGGGGTTTTGTTGCACTACGCTGTCTTGGCGTCCTTCTGTTGGATGCTGGTCGCAGCTATCATTTCGTATAGGAAACTGGTGATAGTGTTCTCCAGAGACGCTTCGCACAAGCTGCTGAAGGCGTCCGCGTTTTCGTGGGGAGCTCCGTGTGCCGTGGTCGGCGTGCTTCTGTCGGTTCGCTCCGCACTCGTACGCGGGACAATTCGACGGAGACGTTCGCAGCGGGGCCTTCTGCTATCCTCGGGGCTGTCGTTGTGGCTGACGGTGTACGCTCCCATCGCCGTGATGCTGCTGGTGAACTGGACGTTATTCCTCCTGATAGTGCGCTCCGTGTTCGCTTCCCGAAGGATCCAGAGGCACGGGGACTCGAACGAGGCCGCTCCGTTGTGCGTCCGTCTCCTGTCTTCTGGTCTTCTTGTTCGGTCTGCCGTGGGTCTTCGGTTTGTTCCTGAACAACATCGTCACGGTGTACTTGTTCAACTTGACCGCGACCTTTCAAGGTTTCGTGCTGTTCCTCTTCTTTGTCCCGGCAATAAGAAGACCAGGGATCTGTGGTTGAACAAGTTGAAGATCAAGCAGACGCGCAAGGTTCCGGTCACGTCCTCGACGTATACGAATAGGAGCACCGGTGGAAACCCTGGCTCCAGGGTAGGGAACACAAGTGTACCTGCGCAATCCAAAGAGACCAAGCCCAGGTCTATGTCTTCCAGTGACGACTCAAGGTTCTCTTGA
Protein
MRGLRLATRDTDAFTDHAFEVIRDEINSSLLQPELSDVVVKIPGSVSVSNRRVSFVVFRTDRAFRNPGSEYSVNSRVVSIKIGNLTRFQNEEYDCNFTDDVNVKSKSVMILGDSIGCSAIKRPTNDCRLFQFIDIYLNPIVELRGSKQKRVCAYWEFLEDGTGYWSQDGCVLMRSEDEGMLDYCRCTHLTHFAEILIPRTSFSDAHENALELISVIGCCVSILGLVLVAITAILFRSWRRDFTNKIWLQLCLAIFITSVCFLVIVFADFLEYNIACMLVGVLLHYAVLASFCWMLVAAIISYRKLVIVFSRDASHKLLKASAFSWGAPCAVVGVLLSVRSALVRGTIRRRRSQRGLLLSSGLSLWLTVYAPIAVMLLVNWTLFLLIVRSVFASRRIQRHGDSNEAAPLCVRLLSSGLLVRSAVGLRFVPEQHRHGVLVQLDRDLSRFRAVPLLCPGNKKTRDLWLNKLKIKQTRKVPVTSSTYTNRSTGGNPGSRVGNTSVPAQSKETKPRSMSSSDDSRFS
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF56436
SSF56436
ProteinModelPortal
Ontologies
GO
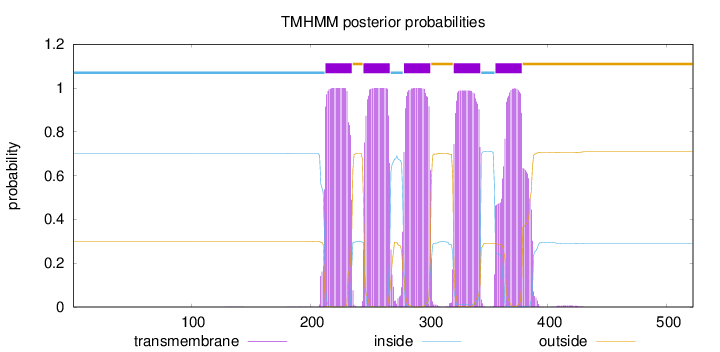
Topology
Length:
522
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.69066
Exp number, first 60 AAs:
0.00032
Total prob of N-in:
0.70180
inside
1 - 212
TMhelix
213 - 235
outside
236 - 244
TMhelix
245 - 267
inside
268 - 278
TMhelix
279 - 301
outside
302 - 320
TMhelix
321 - 343
inside
344 - 355
TMhelix
356 - 378
outside
379 - 522
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
