Pre Gene Modal
BGIBMGA009780
Annotation
PREDICTED:_unconventional_myosin-Va_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.53
Sequence
CDS
ATGTTCGAGTACAGGATACAGGACGAGCCCACCATCATAAAGAAACTTATAACCGATTTGAAACCGAAAGTGGCGACCACGCTGCTGCCGGGCCTGCCCGCCTACATCCTGTTCATGATGCTGCGCCACACGGACCACGTGGACGACGAGCCCAAGATGCACCAGCTCATGAAGGCGGTGCGGAACGGCGTCAAGAAGACCCTGAAGAAGCGATCGGACAGCATCGAGTACAACGCGCTGTGGCTCGCCAACATGCTGAGACTCCTGAACAACCTGCGCCAGTACAGCGGGGACGCGGTGTACCAGGCGGCGAACACTCCGCGCCAGAACCAGCAGTGTCTCCGGATCTTCGACCTCTCGGAGTACCGGCAGGTGCTCAGCGATATAGCCATCTGGATCTACCAGGGTCTGATCAATCTGCTGGAGAGGCAGTTGGACCGTCTGATAGTGCCCGCGATCCTGGAGCACGAGGAGATCAGCGGGCTGTCGGGCCCCGCGCCCCCGCCGCGCCCCCCGCCTCGCCCCGCGCCGGCCCGCGCGCCTCAAGCACGAGCTGCGCTCCGCCTCAGCATCTTCAAGCAGCTCCTGTACTACATCTGCGCGTACAGCTTGAACCAGCTGCTCCTCCGCAAGGACCTGTGCTGCTGGGCTAAGGGCTTGCAGATCAGGTACAACATCTCCCACCTCGAGACATGGATCAAAGAGAACTTGGCCGAATACGGACAGAAAAATGTGGAGGAGGTGTTGAGCGTCTTGAAGCCGATAACCCAGGCCGTGCAGCTGCTGCAGGCCAGAAAGTCCATGCAGGACGTCCAGAGCACCGTGGACATGTGCTCCAACCTGACCGCCATGCAAGTCTGCAAGATTCTGAACATGTACACACCAGCCGAGGAGTACGAGGTGAAGGTGACGCGGGAGTTCATTCACGAGATCCAGAAGAAGATGCAGGAGAAGGCCGGCTCCAATCCGGACAAGGAACCCCAAAACCTACTGATGAACTCCAAGATGATATTCTCCGTACAATTCCCCTTCAACCCCTCACCAATACGACTGGAGGCGGTCGAGCTCCCGGAGATATTGGAGCTAGAGGGCGTACTCACCAAAATATAA
Protein
MFEYRIQDEPTIIKKLITDLKPKVATTLLPGLPAYILFMMLRHTDHVDDEPKMHQLMKAVRNGVKKTLKKRSDSIEYNALWLANMLRLLNNLRQYSGDAVYQAANTPRQNQQCLRIFDLSEYRQVLSDIAIWIYQGLINLLERQLDRLIVPAILEHEEISGLSGPAPPPRPPPRPAPARAPQARAALRLSIFKQLLYYICAYSLNQLLLRKDLCCWAKGLQIRYNISHLETWIKENLAEYGQKNVEEVLSVLKPITQAVQLLQARKSMQDVQSTVDMCSNLTAMQVCKILNMYTPAEEYEVKVTREFIHEIQKKMQEKAGSNPDKEPQNLLMNSKMIFSVQFPFNPSPIRLEAVELPEILELEGVLTKI
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
A0A2H1V2L3
A0A2A4IVB5
A0A437BRT6
A0A194PV37
A0A2H1WQW9
A0A2A4JEB0
+ More
A0A087U050 A0A0N1PIP6 A0A293N4B2 A0A2J7RQB8 A0A023GG04 L7MEL2 A0A131YID1 A0A147BQF9 A0A1E1XK51 D6WQH7 A0A0P4W5R4 A0A0P4WGV2 A0A1E1X7Z1 A0A1B6L2G0 F6K356 A0A0F7VJF0 V5GXV2 A0A1Y1MHK0 A0A0Y0GML1 A0A2T7PSZ8 V4ACD4 A0A1S3JMP1 A0A1S3HBE4 E0VAZ3 T1FR37 E9H736 A0A0P5P9E7 A0A0P5YZB3 A0A0P5LPI2 A0A0P6CNX1 A0A0N8CXS1 A0A0P6EJY1 A0A0P6CWR1 A0A0P5VR14 A0A0P6HK55 A0A0N8CE72 A0A0P6FW99 A0A0P5MQ83 R7T565 A0A0P6BUJ9 E2A5K9 A0A0T6BA34 A0A195BJ13 A0A195DEK4 A0A195FNW2 F4X5I3 A0A151WYN4 A0A195CUA2 E2AL94 A0A026WN41 A0A3L8DE04 E2B2T6 A0A0J7L868 K7IV32 A0A232FB52 A0A2A3E8F5 A0A3N0Z9M1 A0A3P9IV48 A0A452S0B7 A0A093GYL2 A0A2D4LA09 F7AL27 A0A091TQK0 A0A2D4EQH4 A0A091F3U8 A0A091L199 A0A093QHY3 A0A3P9KNU0 A0A093IN05 A0A091HWK8 A0A091FV17 A0A087VMH2 A0A2D4P7X5 A0A091QMI5 A0A452IBV0 A0A091LCF7 A0A093CND2 H2LLF2 A0A087QWM3 A0A091NKI8 A0A091WHL3 A0A091VHS7 A0A091RRU8 U3KBK4 A0A091QM39 A0A2I0MDL5 A0A091M4B0 A0A093PIL1 A0A099ZF97 A0A091R1J4
A0A087U050 A0A0N1PIP6 A0A293N4B2 A0A2J7RQB8 A0A023GG04 L7MEL2 A0A131YID1 A0A147BQF9 A0A1E1XK51 D6WQH7 A0A0P4W5R4 A0A0P4WGV2 A0A1E1X7Z1 A0A1B6L2G0 F6K356 A0A0F7VJF0 V5GXV2 A0A1Y1MHK0 A0A0Y0GML1 A0A2T7PSZ8 V4ACD4 A0A1S3JMP1 A0A1S3HBE4 E0VAZ3 T1FR37 E9H736 A0A0P5P9E7 A0A0P5YZB3 A0A0P5LPI2 A0A0P6CNX1 A0A0N8CXS1 A0A0P6EJY1 A0A0P6CWR1 A0A0P5VR14 A0A0P6HK55 A0A0N8CE72 A0A0P6FW99 A0A0P5MQ83 R7T565 A0A0P6BUJ9 E2A5K9 A0A0T6BA34 A0A195BJ13 A0A195DEK4 A0A195FNW2 F4X5I3 A0A151WYN4 A0A195CUA2 E2AL94 A0A026WN41 A0A3L8DE04 E2B2T6 A0A0J7L868 K7IV32 A0A232FB52 A0A2A3E8F5 A0A3N0Z9M1 A0A3P9IV48 A0A452S0B7 A0A093GYL2 A0A2D4LA09 F7AL27 A0A091TQK0 A0A2D4EQH4 A0A091F3U8 A0A091L199 A0A093QHY3 A0A3P9KNU0 A0A093IN05 A0A091HWK8 A0A091FV17 A0A087VMH2 A0A2D4P7X5 A0A091QMI5 A0A452IBV0 A0A091LCF7 A0A093CND2 H2LLF2 A0A087QWM3 A0A091NKI8 A0A091WHL3 A0A091VHS7 A0A091RRU8 U3KBK4 A0A091QM39 A0A2I0MDL5 A0A091M4B0 A0A093PIL1 A0A099ZF97 A0A091R1J4
Pubmed
EMBL
ODYU01000388
SOQ35087.1
NWSH01006433
PCG63436.1
RSAL01000015
RVE53162.1
+ More
KQ459593 KPI96619.1 ODYU01010387 SOQ55465.1 NWSH01001891 PCG69780.1 KK117539 KFM70739.1 KQ460280 KPJ16326.1 GFWV01023237 MAA47964.1 NEVH01001344 PNF43034.1 GBBM01003505 JAC31913.1 GACK01002599 JAA62435.1 GEDV01009513 JAP79044.1 GEGO01002425 JAR92979.1 GFAA01003816 JAT99618.1 KQ971354 EFA06975.2 GDRN01089957 JAI60556.1 GDRN01089955 JAI60557.1 GFAC01003801 JAT95387.1 GEBQ01022099 JAT17878.1 HM212671 ADM15669.1 LN830727 CFW94236.1 GALX01003268 JAB65198.1 GEZM01031022 JAV85121.1 KU193727 AMB26737.1 PZQS01000002 PVD36507.1 KB202284 ESO90956.1 DS235019 EEB10549.1 AMQM01001739 KB097639 ESN92553.1 GL732599 EFX72449.1 GDIQ01141357 JAL10369.1 GDIP01052936 JAM50779.1 GDIQ01170346 GDIQ01136811 GDIQ01030299 LRGB01003163 JAK81379.1 KZS03930.1 GDIQ01090022 JAN04715.1 GDIP01088479 JAM15236.1 GDIQ01061683 JAN33054.1 GDIQ01086333 JAN08404.1 GDIP01096726 JAM06989.1 GDIQ01018624 JAN76113.1 GDIP01140471 JAL63243.1 GDIQ01041893 JAN52844.1 GDIQ01152952 JAK98773.1 AMQN01015508 KB311976 ELT88116.1 GDIP01017260 JAM86455.1 GL436993 EFN71292.1 LJIG01002798 KRT84142.1 KQ976464 KYM84426.1 KQ980934 KYN11323.1 KQ981382 KYN42143.1 GL888719 EGI58273.1 KQ982652 KYQ52811.1 KQ977279 KYN04261.1 GL440553 EFN65790.1 KK107148 EZA57445.1 QOIP01000009 RLU18383.1 GL445235 EFN90001.1 LBMM01000310 KMR01422.1 AAZX01001194 NNAY01000532 OXU27842.1 KZ288326 PBC27980.1 RJVU01006614 ROL54648.1 KL216904 KFV71897.1 IACL01118327 LAB17673.1 KK460780 KFQ79009.1 IACJ01012249 LAA37508.1 KK719540 KFO63936.1 KK550905 KFP33739.1 KL671826 KFW83662.1 KK591087 KFW00159.1 KL217820 KFO99512.1 KL447385 KFO72801.1 KL498708 KFO13814.1 IACN01056768 LAB54072.1 KK702820 KFQ28398.1 KL316499 KFP54054.1 KL462157 KFV13812.1 KL225960 KFM05627.1 KK836012 KFP80000.1 KK735633 KFR15177.1 KL411013 KFR02679.1 KK931736 KFQ45385.1 AGTO01000576 KK664707 KFQ10524.1 AKCR02000019 PKK27767.1 KK520484 KFP66231.1 KL225418 KFW72027.1 KL892401 KGL79708.1 KK807436 KFQ33099.1
KQ459593 KPI96619.1 ODYU01010387 SOQ55465.1 NWSH01001891 PCG69780.1 KK117539 KFM70739.1 KQ460280 KPJ16326.1 GFWV01023237 MAA47964.1 NEVH01001344 PNF43034.1 GBBM01003505 JAC31913.1 GACK01002599 JAA62435.1 GEDV01009513 JAP79044.1 GEGO01002425 JAR92979.1 GFAA01003816 JAT99618.1 KQ971354 EFA06975.2 GDRN01089957 JAI60556.1 GDRN01089955 JAI60557.1 GFAC01003801 JAT95387.1 GEBQ01022099 JAT17878.1 HM212671 ADM15669.1 LN830727 CFW94236.1 GALX01003268 JAB65198.1 GEZM01031022 JAV85121.1 KU193727 AMB26737.1 PZQS01000002 PVD36507.1 KB202284 ESO90956.1 DS235019 EEB10549.1 AMQM01001739 KB097639 ESN92553.1 GL732599 EFX72449.1 GDIQ01141357 JAL10369.1 GDIP01052936 JAM50779.1 GDIQ01170346 GDIQ01136811 GDIQ01030299 LRGB01003163 JAK81379.1 KZS03930.1 GDIQ01090022 JAN04715.1 GDIP01088479 JAM15236.1 GDIQ01061683 JAN33054.1 GDIQ01086333 JAN08404.1 GDIP01096726 JAM06989.1 GDIQ01018624 JAN76113.1 GDIP01140471 JAL63243.1 GDIQ01041893 JAN52844.1 GDIQ01152952 JAK98773.1 AMQN01015508 KB311976 ELT88116.1 GDIP01017260 JAM86455.1 GL436993 EFN71292.1 LJIG01002798 KRT84142.1 KQ976464 KYM84426.1 KQ980934 KYN11323.1 KQ981382 KYN42143.1 GL888719 EGI58273.1 KQ982652 KYQ52811.1 KQ977279 KYN04261.1 GL440553 EFN65790.1 KK107148 EZA57445.1 QOIP01000009 RLU18383.1 GL445235 EFN90001.1 LBMM01000310 KMR01422.1 AAZX01001194 NNAY01000532 OXU27842.1 KZ288326 PBC27980.1 RJVU01006614 ROL54648.1 KL216904 KFV71897.1 IACL01118327 LAB17673.1 KK460780 KFQ79009.1 IACJ01012249 LAA37508.1 KK719540 KFO63936.1 KK550905 KFP33739.1 KL671826 KFW83662.1 KK591087 KFW00159.1 KL217820 KFO99512.1 KL447385 KFO72801.1 KL498708 KFO13814.1 IACN01056768 LAB54072.1 KK702820 KFQ28398.1 KL316499 KFP54054.1 KL462157 KFV13812.1 KL225960 KFM05627.1 KK836012 KFP80000.1 KK735633 KFR15177.1 KL411013 KFR02679.1 KK931736 KFQ45385.1 AGTO01000576 KK664707 KFQ10524.1 AKCR02000019 PKK27767.1 KK520484 KFP66231.1 KL225418 KFW72027.1 KL892401 KGL79708.1 KK807436 KFQ33099.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000054359
UP000053240
UP000235965
+ More
UP000007266 UP000245119 UP000030746 UP000085678 UP000009046 UP000015101 UP000000305 UP000076858 UP000014760 UP000000311 UP000078540 UP000078492 UP000078541 UP000007755 UP000075809 UP000078542 UP000053097 UP000279307 UP000008237 UP000036403 UP000002358 UP000215335 UP000242457 UP000265200 UP000291022 UP000053875 UP000002281 UP000052976 UP000053258 UP000265180 UP000054308 UP000053760 UP000291020 UP000001038 UP000053286 UP000053605 UP000053283 UP000016665 UP000053872 UP000054081 UP000053641
UP000007266 UP000245119 UP000030746 UP000085678 UP000009046 UP000015101 UP000000305 UP000076858 UP000014760 UP000000311 UP000078540 UP000078492 UP000078541 UP000007755 UP000075809 UP000078542 UP000053097 UP000279307 UP000008237 UP000036403 UP000002358 UP000215335 UP000242457 UP000265200 UP000291022 UP000053875 UP000002281 UP000052976 UP000053258 UP000265180 UP000054308 UP000053760 UP000291020 UP000001038 UP000053286 UP000053605 UP000053283 UP000016665 UP000053872 UP000054081 UP000053641
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2H1V2L3
A0A2A4IVB5
A0A437BRT6
A0A194PV37
A0A2H1WQW9
A0A2A4JEB0
+ More
A0A087U050 A0A0N1PIP6 A0A293N4B2 A0A2J7RQB8 A0A023GG04 L7MEL2 A0A131YID1 A0A147BQF9 A0A1E1XK51 D6WQH7 A0A0P4W5R4 A0A0P4WGV2 A0A1E1X7Z1 A0A1B6L2G0 F6K356 A0A0F7VJF0 V5GXV2 A0A1Y1MHK0 A0A0Y0GML1 A0A2T7PSZ8 V4ACD4 A0A1S3JMP1 A0A1S3HBE4 E0VAZ3 T1FR37 E9H736 A0A0P5P9E7 A0A0P5YZB3 A0A0P5LPI2 A0A0P6CNX1 A0A0N8CXS1 A0A0P6EJY1 A0A0P6CWR1 A0A0P5VR14 A0A0P6HK55 A0A0N8CE72 A0A0P6FW99 A0A0P5MQ83 R7T565 A0A0P6BUJ9 E2A5K9 A0A0T6BA34 A0A195BJ13 A0A195DEK4 A0A195FNW2 F4X5I3 A0A151WYN4 A0A195CUA2 E2AL94 A0A026WN41 A0A3L8DE04 E2B2T6 A0A0J7L868 K7IV32 A0A232FB52 A0A2A3E8F5 A0A3N0Z9M1 A0A3P9IV48 A0A452S0B7 A0A093GYL2 A0A2D4LA09 F7AL27 A0A091TQK0 A0A2D4EQH4 A0A091F3U8 A0A091L199 A0A093QHY3 A0A3P9KNU0 A0A093IN05 A0A091HWK8 A0A091FV17 A0A087VMH2 A0A2D4P7X5 A0A091QMI5 A0A452IBV0 A0A091LCF7 A0A093CND2 H2LLF2 A0A087QWM3 A0A091NKI8 A0A091WHL3 A0A091VHS7 A0A091RRU8 U3KBK4 A0A091QM39 A0A2I0MDL5 A0A091M4B0 A0A093PIL1 A0A099ZF97 A0A091R1J4
A0A087U050 A0A0N1PIP6 A0A293N4B2 A0A2J7RQB8 A0A023GG04 L7MEL2 A0A131YID1 A0A147BQF9 A0A1E1XK51 D6WQH7 A0A0P4W5R4 A0A0P4WGV2 A0A1E1X7Z1 A0A1B6L2G0 F6K356 A0A0F7VJF0 V5GXV2 A0A1Y1MHK0 A0A0Y0GML1 A0A2T7PSZ8 V4ACD4 A0A1S3JMP1 A0A1S3HBE4 E0VAZ3 T1FR37 E9H736 A0A0P5P9E7 A0A0P5YZB3 A0A0P5LPI2 A0A0P6CNX1 A0A0N8CXS1 A0A0P6EJY1 A0A0P6CWR1 A0A0P5VR14 A0A0P6HK55 A0A0N8CE72 A0A0P6FW99 A0A0P5MQ83 R7T565 A0A0P6BUJ9 E2A5K9 A0A0T6BA34 A0A195BJ13 A0A195DEK4 A0A195FNW2 F4X5I3 A0A151WYN4 A0A195CUA2 E2AL94 A0A026WN41 A0A3L8DE04 E2B2T6 A0A0J7L868 K7IV32 A0A232FB52 A0A2A3E8F5 A0A3N0Z9M1 A0A3P9IV48 A0A452S0B7 A0A093GYL2 A0A2D4LA09 F7AL27 A0A091TQK0 A0A2D4EQH4 A0A091F3U8 A0A091L199 A0A093QHY3 A0A3P9KNU0 A0A093IN05 A0A091HWK8 A0A091FV17 A0A087VMH2 A0A2D4P7X5 A0A091QMI5 A0A452IBV0 A0A091LCF7 A0A093CND2 H2LLF2 A0A087QWM3 A0A091NKI8 A0A091WHL3 A0A091VHS7 A0A091RRU8 U3KBK4 A0A091QM39 A0A2I0MDL5 A0A091M4B0 A0A093PIL1 A0A099ZF97 A0A091R1J4
PDB
4LNZ
E-value=2.54971e-74,
Score=708
Ontologies
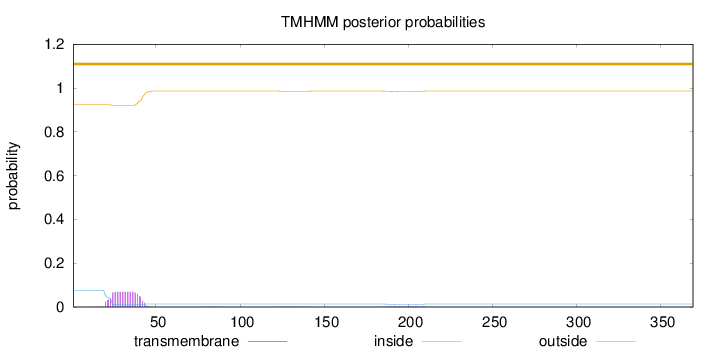
Topology
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.46306
Exp number, first 60 AAs:
1.38176
Total prob of N-in:
0.07615
outside
1 - 369
Population Genetic Test Statistics
Pi
1.115362
Theta
2.825993
Tajima's D
-2.006239
CLR
0
CSRT
0.00764961751912404
Interpretation
Uncertain
