Gene
KWMTBOMO16680
Pre Gene Modal
BGIBMGA014553
Annotation
hypothetical_protein_TcasGA2_TC034914?_partial_[Tribolium_castaneum]
Location in the cell
Cytoplasmic Reliability : 1.522 Nuclear Reliability : 2.07
Sequence
CDS
ATGTTCCATGATGCGACCAAACAGGACGGCCGAAGTCAAAAGGACATGTATGAGCTTCTGTCAGAATTAACCAAAATAGTGGAAGAGATGGGAAACAAAGAGCTGACGATTATAAAGCCCCAAGGAATCGATACAGGTGTAACAAGGAAGATGTTAGAAGTACTGTTCCGTGGAAAGGACATCAAACTAGAATTAAGGACACCTAAAAAAGGAGAAAATACAAACAATACGGAAACAAGAAAACGGGACGATGTAATAGTCATAGAAGGAGGGACAACTACTTATGCGGACACCGTAAAGAGGCTAAAAGAAGGAATAAAAGGGACCGCATCCCTAGAAAACATAAAAGGTGTAAAAAAGACTGCCAAGGGACAAGTACTGGTAAGAGTGATGGGAAATATTGAGGAGATGACTGGGAAAATAAGAGAAATGATGGGAGACAGAAAGGTAAGAAGAACTGGGGGGAGCAAGAAAAAAGTATTACACATAAAGGGAATGGACAGCTCGGTGGAAATGGAGGAAGTAAGGGAGGACTTAAAGTCGACAAACTTGATAAAAAAAGTAGACAGCTTGGAGATAAGATCACTGAGACCGATGGGCGGTGGAAACCAGACGGCAACAGTGATCCTGGATGAAGAGGATGCAGGAAGGCTGCTTGCGGGTGGGAAGATAAGAGTAGGGCTAAATATATGCCACATTCATGAAAGAATTGAACTGCTGAGATGCTACCGGTGCTGGGAATATGGCCACAAAGCGGAAAACTGCAACGTAAAAAAGGACACACAGCTGGCACCGGTAGATGCCCACTCTTCCGGGAGGCATTGGAAACCGCGAAGCGAAGTCAAACCTATAACAAAAATGGTTTTTAAAGTGTTACAGATAAATATGGACCGAGGGAAAGCCGCGCACGACCTACTTCTGGCAACTGCGAGCAAAATGAAAGCGGATCTCGTGATGCTAGCTGAACCCAATAGAAAGCTGGCAAAAGCTGCTGGATGGGAGTGCGACACCAGGGGAGATGCAGCCCTAGTGATTCTTAATAAAACACTGACGGTTTCTGAAATTTATAGGGGGAAGGGATTTGTTGGGTTCAAGATGAAGCACTTTGCCGCGTTCAGCTGTTACATATCTCCCAACTGCAGTTTTGAGGAGTTCAAAGCTGACGTCGATGAGCTTTGGACACTACTTAAAAAACAAAAGACACCGATCCTAGTGGCAGGAGATTTTAACTCTGCATCGAGAGCGTGGGGCAGTAAAGTAACCAACAAAAGGGGCCACTACCTGCTGGAAATGGTGGCGGACCTCGACACTGTCATCATGAATGAAGGCGTGACCCCAACTTTTGAAAGAGGAGACAGAACATCCAGGCCCGACATCACCATGGCCTCTGAAACGCTGGCAGGCAAGATCAGTAATTGGCAAGTGCTCGACGAGGAGACCATGAGCGGACATAAATATATAGCCTACGAGATAAAGCATGTATTGCCTCTATCCCAGTGCCATGCACAGATGAAAAGGTGGAACCTCCAAAGGCTGGACGTAGATAAGTTCACTAAAAGCATGAAAGGAGCGCAGATCTCGACTCCCATGGAACTGGTGCAGGCTACGACGGCAGCGTGTGACGCTTCTATGCCAAAGCAAAACAACCGCAAGAAGAAAGGAGTCTACTGGTGGAACAAGGAAATTGCGGAAGCAAGGAAGGAGTGTTTGAAATGCCGAAGGAGGTACACAAGGAATAACAAAAAGCTAGGAAAAGGGAAAGACAGGGAAAGTAACGAAGAAAATGAACACAAAAAGCAGTATAAGGAAGCGAAACTGGAGCTACAAAGACGAATATGGAAAGCGAAAGACGATAAATGGAGGGAAGTGTGCGAGGAAGTCGACAATGATATATGGGGACTGGGATACAAAATAGTGTCAAAAAAACTACGCGGAAACTCACCGATAAATATGGGCATGGCCCAAATTCAAAGCATAGCTGATGGCCTCTTCCCGGAACACATAGATTTCGAACGGAAAACGAAACCATCACAGGAGTTCCCAGAGGTGACCGAGGAGGAGATCATCCAACTAAAAGCTCGCATAAAGCTCAGGAAAGCCCCAGGGCCCGACCGGATTCCGCCGGAGATAGTGACTCTAGCAGTAGCAGTAATTCCAGACAGGATAAGGCTTGTGTTGCAGAAGGTGCTGGAGTCTGGCGTCTTCCCCAACAGCTGGAAAACAGCCAGACTAGTCCTGCTAAGAAAACCAGGCAAACCCGCTGAAGATCCTGCGGGGCACAGGCCGCTATGTCTACTGGACTGCTACGGAAAGCTGCTGGAGTCGCTGATCCTAGGCAGAATAGAAAAACAACTAGTGCAATCGGACTGTTTACAAGATTCACAGTACGGGTTCAGAAAGGGGAGGTCCACAGTGGATGCAGCCAACAAGCTACTTGAGATAGCGGAAGACGCGAGACGAGGAACCCACCGCACCAGAAGAATCTGCGCAGTAGTGGCCCTCGACGTCCGAAATGCTTTCAATTCTGCATCGTGGGAGCTCATCCTGGAGGAAATGGCTGCAATAGGTATGCCTCAATACATCTATAATATCATAAACAGCTACCTCCAGGACAGATGCATCATACTGACAGACGCCGAGGGAGGGGCTGTAGAGAAAAAGGTTACCGGTGGTGTTCCGCAGGGGTCGATACTGGGCCCCACGCTGTGGAACATACTCTATGATGGGGTACTGCGCCTCGAACTGGGGGACGGGGCAAAATTAATAGGCTTTGCTGACGATTTAGCTCTGGTGGTGTCAGCAAAGAAGGAAGCAGAACTGATGTCAATAACGAACGTAGCTCTAGAAAAGATCAGCGCTTGGATGAAGCAGAAGCGGCTACACTTGGCCCCAGAAAAGACCGAGGCAATTGTCTTAAGTGGGCGAAGGAAGCTGAGCGAGATCTGCTTTACCCTCCAGGGCGTCAGGGTGGTTCCAACCGACAAACTTAAATACCTTGGGTTGTGGTTTGACAAAAGTTTGAAGTTCGGTAAACACATTGAGGAAACAACGGGTAAAGCCGAAAGATTGACGGCCGCCCTTGGCCAAATTATGCCCAGAAAAGGAGGGCCGAGGTCAAGCAAGCGCAGACTACTGGCATCCGCGATACAATCTGTGATGTTATATGGAGCACCGGTGTGGGCAAGGGTAATGAATGTAGCGAAGTACAGGAACATGTATAGCAAAGTGCAACGACAGCTGGCAATACGCATCTGTGGAGCTTACCGTACTATATCGCTAGACGCCGTCTTGGTGATAGCGGGCCTGATACCGATAGAACGGCTAGCAAAGGAACGGCAACGCTACTACCACGAGGGGACAAGAACGAGAGAAGAGGAAAGAGCACATACAATAAAAGAATGGGAACGAGACTGGCAGAAGGAGGGAAAAGGTAAGTGGACTCGTACAATCATACCCAACCTTAGGCTCTGGATGGGACGGAAACATGGCGAAACCGACAGACTGCTGACACAGGCCCTAAGTGGGCATGGAGTTTTCAACACTTACCTGCATGCAATAGGGAAAGCAGAAAGCGCAAGTTGTCCATACTGCGAACATGTTGACACACCGGAACATGCCCTATTTCAATGCAAAAAGTTCTCAGAACTAAGAGAGAAAACGGAGGGAAGTGACGGAAAGGTGACTGTGGAAAACCTGATGGTGAGAATGATCCATAGTAAAGAAGAATGGGATAAATATGCGGGAATGCTCAAAGAGATAATGAGAGTGCGGGAAAAGGACGAAAGACTAAGGCAAGAATCTATCTATAATAATATATAA
Protein
MFHDATKQDGRSQKDMYELLSELTKIVEEMGNKELTIIKPQGIDTGVTRKMLEVLFRGKDIKLELRTPKKGENTNNTETRKRDDVIVIEGGTTTYADTVKRLKEGIKGTASLENIKGVKKTAKGQVLVRVMGNIEEMTGKIREMMGDRKVRRTGGSKKKVLHIKGMDSSVEMEEVREDLKSTNLIKKVDSLEIRSLRPMGGGNQTATVILDEEDAGRLLAGGKIRVGLNICHIHERIELLRCYRCWEYGHKAENCNVKKDTQLAPVDAHSSGRHWKPRSEVKPITKMVFKVLQINMDRGKAAHDLLLATASKMKADLVMLAEPNRKLAKAAGWECDTRGDAALVILNKTLTVSEIYRGKGFVGFKMKHFAAFSCYISPNCSFEEFKADVDELWTLLKKQKTPILVAGDFNSASRAWGSKVTNKRGHYLLEMVADLDTVIMNEGVTPTFERGDRTSRPDITMASETLAGKISNWQVLDEETMSGHKYIAYEIKHVLPLSQCHAQMKRWNLQRLDVDKFTKSMKGAQISTPMELVQATTAACDASMPKQNNRKKKGVYWWNKEIAEARKECLKCRRRYTRNNKKLGKGKDRESNEENEHKKQYKEAKLELQRRIWKAKDDKWREVCEEVDNDIWGLGYKIVSKKLRGNSPINMGMAQIQSIADGLFPEHIDFERKTKPSQEFPEVTEEEIIQLKARIKLRKAPGPDRIPPEIVTLAVAVIPDRIRLVLQKVLESGVFPNSWKTARLVLLRKPGKPAEDPAGHRPLCLLDCYGKLLESLILGRIEKQLVQSDCLQDSQYGFRKGRSTVDAANKLLEIAEDARRGTHRTRRICAVVALDVRNAFNSASWELILEEMAAIGMPQYIYNIINSYLQDRCIILTDAEGGAVEKKVTGGVPQGSILGPTLWNILYDGVLRLELGDGAKLIGFADDLALVVSAKKEAELMSITNVALEKISAWMKQKRLHLAPEKTEAIVLSGRRKLSEICFTLQGVRVVPTDKLKYLGLWFDKSLKFGKHIEETTGKAERLTAALGQIMPRKGGPRSSKRRLLASAIQSVMLYGAPVWARVMNVAKYRNMYSKVQRQLAIRICGAYRTISLDAVLVIAGLIPIERLAKERQRYYHEGTRTREEERAHTIKEWERDWQKEGKGKWTRTIIPNLRLWMGRKHGETDRLLTQALSGHGVFNTYLHAIGKAESASCPYCEHVDTPEHALFQCKKFSELREKTEGSDGKVTVENLMVRMIHSKEEWDKYAGMLKEIMRVREKDERLRQESIYNNI
Summary
Uniprot
A0A139WAH4
A0A139WNC9
A0A139WN93
A0A139W940
J9KJG8
A0A139W9B3
+ More
A0A139W939 F7IYV5 A0A139W9A6 F7IYU8 F7IYV2 D6WC19 V5I881 A0A2H8TR36 A0A139W9J0 J9JWJ3 X1WZ64 J9KM84 T1DCM0 A0A224XIZ2 T1DG89 T1DG59 A0A142LX45 F7IYV6 A0A2M4CS98 J9LVU7 J9KUM6 A0A2M4ADW9 A0A2S2NQQ6 A0A139W942 A0A034WR48 A0A2M4BBV9 A0A2M4BC28 A0A2M4BC24 A0A2M4BC01 A0A2M4BC18 J9K6F0 A0A142LX43 W8ADE4 W8ANK7 A0A2H8TIE5 J9KNS1 A0A142LX37 F7IYV3 A0A0K8VPH1 A0A2M4CSA9 J9JY94 Q868S4 A0A2M4BDI3 A0A2M4CSI6 F7IYV7 J9KT61 A0A2S2PRL8 A0A2M4BB78 A0A2M4BB89 A0A2M3Z1C1 X1WJY4 D7EIB0 Q868S6 X1WXT5 A0A2M4BBA0 A0A139WA31 A0A2S2PEZ7 J9LA43 A0A2M4AK35 Q868S0 A0A139W8J5 Q9N9Z1 J9L0J0 Q868T0 A0A2M4BCE1 J9LQQ1 J9LM37 Q868S8 V9GZD3 A0A2M4CJ33 J9KNN8 Q868R6 A0A2S2P7D1 Q868R2 Q868R4 A0A0J7KIY5 Q868R0 J9LBG0 J9M6A5 Q868Q8
A0A139W939 F7IYV5 A0A139W9A6 F7IYU8 F7IYV2 D6WC19 V5I881 A0A2H8TR36 A0A139W9J0 J9JWJ3 X1WZ64 J9KM84 T1DCM0 A0A224XIZ2 T1DG89 T1DG59 A0A142LX45 F7IYV6 A0A2M4CS98 J9LVU7 J9KUM6 A0A2M4ADW9 A0A2S2NQQ6 A0A139W942 A0A034WR48 A0A2M4BBV9 A0A2M4BC28 A0A2M4BC24 A0A2M4BC01 A0A2M4BC18 J9K6F0 A0A142LX43 W8ADE4 W8ANK7 A0A2H8TIE5 J9KNS1 A0A142LX37 F7IYV3 A0A0K8VPH1 A0A2M4CSA9 J9JY94 Q868S4 A0A2M4BDI3 A0A2M4CSI6 F7IYV7 J9KT61 A0A2S2PRL8 A0A2M4BB78 A0A2M4BB89 A0A2M3Z1C1 X1WJY4 D7EIB0 Q868S6 X1WXT5 A0A2M4BBA0 A0A139WA31 A0A2S2PEZ7 J9LA43 A0A2M4AK35 Q868S0 A0A139W8J5 Q9N9Z1 J9L0J0 Q868T0 A0A2M4BCE1 J9LQQ1 J9LM37 Q868S8 V9GZD3 A0A2M4CJ33 J9KNN8 Q868R6 A0A2S2P7D1 Q868R2 Q868R4 A0A0J7KIY5 Q868R0 J9LBG0 J9M6A5 Q868Q8
EMBL
KQ971388
KYB24913.1
KQ971310
KYB29500.1
KYB29499.1
KQ972691
+ More
KXZ75806.1 ABLF02041886 KQ972544 KXZ75869.1 KXZ75807.1 AB593325 BAK38646.1 KXZ75870.1 AB593321 BAK38639.1 AB593323 BAK38643.1 KQ971309 EEZ99146.1 GALX01005145 JAB63321.1 GFXV01003903 MBW15708.1 KQ972226 KYB24577.1 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 GALA01001777 JAA93075.1 GFTR01008427 JAW07999.1 GALA01001776 JAA93076.1 GALA01000302 JAA94550.1 KU543683 AMS38371.1 AB593326 BAK38647.1 GGFL01004044 MBW68222.1 ABLF02034925 ABLF02041557 GGFK01005660 MBW38981.1 GGMR01006838 MBY19457.1 KXZ75805.1 GAKP01002292 JAC56660.1 GGFJ01001369 MBW50510.1 GGFJ01001439 MBW50580.1 GGFJ01001454 MBW50595.1 GGFJ01001370 MBW50511.1 GGFJ01001453 MBW50594.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 KU543682 AMS38369.1 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GFXV01002090 MBW13895.1 ABLF02018808 KU543679 AMS38363.1 AB593324 BAK38644.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 GGFL01004046 MBW68224.1 ABLF02007989 AB090815 BAC57906.1 GGFJ01001727 MBW50868.1 GGFL01004047 MBW68225.1 AB593327 BAK38648.1 ABLF02009073 ABLF02030702 ABLF02042963 GGMR01019493 MBY32112.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 GGFM01001554 MBW22305.1 ABLF02011238 AB593322 BAK38641.1 AB090814 BAC57904.1 ABLF02004854 ABLF02041317 GGFJ01001166 MBW50307.1 KQ971675 KYB24771.1 GGMR01015159 MBY27778.1 ABLF02018098 GGFK01007771 MBW41092.1 AB090817 BAC57910.1 KQ973256 KXZ75595.1 AJ278684 CAB99192.1 ABLF02027248 AB090812 BAC57900.1 GGFJ01001541 MBW50682.1 ABLF02030663 ABLF02041312 ABLF02014650 AB090813 BAC57902.1 M93690 AAA29363.1 GGFL01001192 MBW65370.1 ABLF02023257 AB090819 BAC57914.1 GGMR01012748 MBY25367.1 AB090821 BAC57918.1 AB090820 BAC57916.1 LBMM01006954 KMQ90166.1 AB090822 BAC57920.1 ABLF02021645 ABLF02020969 AB090823 BAC57922.1
KXZ75806.1 ABLF02041886 KQ972544 KXZ75869.1 KXZ75807.1 AB593325 BAK38646.1 KXZ75870.1 AB593321 BAK38639.1 AB593323 BAK38643.1 KQ971309 EEZ99146.1 GALX01005145 JAB63321.1 GFXV01003903 MBW15708.1 KQ972226 KYB24577.1 ABLF02013358 ABLF02013361 ABLF02054869 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 GALA01001777 JAA93075.1 GFTR01008427 JAW07999.1 GALA01001776 JAA93076.1 GALA01000302 JAA94550.1 KU543683 AMS38371.1 AB593326 BAK38647.1 GGFL01004044 MBW68222.1 ABLF02034925 ABLF02041557 GGFK01005660 MBW38981.1 GGMR01006838 MBY19457.1 KXZ75805.1 GAKP01002292 JAC56660.1 GGFJ01001369 MBW50510.1 GGFJ01001439 MBW50580.1 GGFJ01001454 MBW50595.1 GGFJ01001370 MBW50511.1 GGFJ01001453 MBW50594.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 KU543682 AMS38369.1 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GFXV01002090 MBW13895.1 ABLF02018808 KU543679 AMS38363.1 AB593324 BAK38644.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 GGFL01004046 MBW68224.1 ABLF02007989 AB090815 BAC57906.1 GGFJ01001727 MBW50868.1 GGFL01004047 MBW68225.1 AB593327 BAK38648.1 ABLF02009073 ABLF02030702 ABLF02042963 GGMR01019493 MBY32112.1 GGFJ01001164 MBW50305.1 GGFJ01001165 MBW50306.1 GGFM01001554 MBW22305.1 ABLF02011238 AB593322 BAK38641.1 AB090814 BAC57904.1 ABLF02004854 ABLF02041317 GGFJ01001166 MBW50307.1 KQ971675 KYB24771.1 GGMR01015159 MBY27778.1 ABLF02018098 GGFK01007771 MBW41092.1 AB090817 BAC57910.1 KQ973256 KXZ75595.1 AJ278684 CAB99192.1 ABLF02027248 AB090812 BAC57900.1 GGFJ01001541 MBW50682.1 ABLF02030663 ABLF02041312 ABLF02014650 AB090813 BAC57902.1 M93690 AAA29363.1 GGFL01001192 MBW65370.1 ABLF02023257 AB090819 BAC57914.1 GGMR01012748 MBY25367.1 AB090821 BAC57918.1 AB090820 BAC57916.1 LBMM01006954 KMQ90166.1 AB090822 BAC57920.1 ABLF02021645 ABLF02020969 AB090823 BAC57922.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000663 Natr_peptide
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR021896 Transposase_37
IPR026960 RVT-Znf
IPR016067 S-AdoMet_deCO2ase_core
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000663 Natr_peptide
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR021896 Transposase_37
IPR026960 RVT-Znf
IPR016067 S-AdoMet_deCO2ase_core
Gene 3D
ProteinModelPortal
A0A139WAH4
A0A139WNC9
A0A139WN93
A0A139W940
J9KJG8
A0A139W9B3
+ More
A0A139W939 F7IYV5 A0A139W9A6 F7IYU8 F7IYV2 D6WC19 V5I881 A0A2H8TR36 A0A139W9J0 J9JWJ3 X1WZ64 J9KM84 T1DCM0 A0A224XIZ2 T1DG89 T1DG59 A0A142LX45 F7IYV6 A0A2M4CS98 J9LVU7 J9KUM6 A0A2M4ADW9 A0A2S2NQQ6 A0A139W942 A0A034WR48 A0A2M4BBV9 A0A2M4BC28 A0A2M4BC24 A0A2M4BC01 A0A2M4BC18 J9K6F0 A0A142LX43 W8ADE4 W8ANK7 A0A2H8TIE5 J9KNS1 A0A142LX37 F7IYV3 A0A0K8VPH1 A0A2M4CSA9 J9JY94 Q868S4 A0A2M4BDI3 A0A2M4CSI6 F7IYV7 J9KT61 A0A2S2PRL8 A0A2M4BB78 A0A2M4BB89 A0A2M3Z1C1 X1WJY4 D7EIB0 Q868S6 X1WXT5 A0A2M4BBA0 A0A139WA31 A0A2S2PEZ7 J9LA43 A0A2M4AK35 Q868S0 A0A139W8J5 Q9N9Z1 J9L0J0 Q868T0 A0A2M4BCE1 J9LQQ1 J9LM37 Q868S8 V9GZD3 A0A2M4CJ33 J9KNN8 Q868R6 A0A2S2P7D1 Q868R2 Q868R4 A0A0J7KIY5 Q868R0 J9LBG0 J9M6A5 Q868Q8
A0A139W939 F7IYV5 A0A139W9A6 F7IYU8 F7IYV2 D6WC19 V5I881 A0A2H8TR36 A0A139W9J0 J9JWJ3 X1WZ64 J9KM84 T1DCM0 A0A224XIZ2 T1DG89 T1DG59 A0A142LX45 F7IYV6 A0A2M4CS98 J9LVU7 J9KUM6 A0A2M4ADW9 A0A2S2NQQ6 A0A139W942 A0A034WR48 A0A2M4BBV9 A0A2M4BC28 A0A2M4BC24 A0A2M4BC01 A0A2M4BC18 J9K6F0 A0A142LX43 W8ADE4 W8ANK7 A0A2H8TIE5 J9KNS1 A0A142LX37 F7IYV3 A0A0K8VPH1 A0A2M4CSA9 J9JY94 Q868S4 A0A2M4BDI3 A0A2M4CSI6 F7IYV7 J9KT61 A0A2S2PRL8 A0A2M4BB78 A0A2M4BB89 A0A2M3Z1C1 X1WJY4 D7EIB0 Q868S6 X1WXT5 A0A2M4BBA0 A0A139WA31 A0A2S2PEZ7 J9LA43 A0A2M4AK35 Q868S0 A0A139W8J5 Q9N9Z1 J9L0J0 Q868T0 A0A2M4BCE1 J9LQQ1 J9LM37 Q868S8 V9GZD3 A0A2M4CJ33 J9KNN8 Q868R6 A0A2S2P7D1 Q868R2 Q868R4 A0A0J7KIY5 Q868R0 J9LBG0 J9M6A5 Q868Q8
PDB
1WDU
E-value=5.01964e-13,
Score=185
Ontologies
KEGG
GO
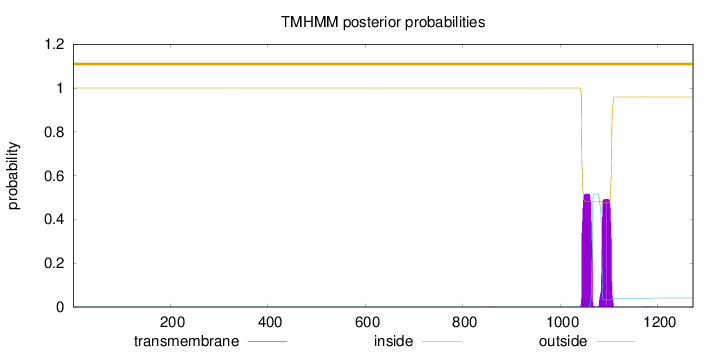
Topology
Length:
1272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
21.15526
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1272
Population Genetic Test Statistics
Pi
3.484797
Theta
5.990565
Tajima's D
-1.590359
CLR
0.009334
CSRT
0.0450477476126194
Interpretation
Uncertain
