Pre Gene Modal
BGIBMGA014079
Annotation
PREDICTED:_BAG_family_molecular_chaperone_regulator_2-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.872
Sequence
CDS
ATGAGACTGATAGCAGTACTAGATCAAGTCGAGATGCGAGTGGAGAGGCTCCGAAAAGACACAGTCAGGATTGAAGAAGAAAAGGACTCCTTACTTTCAACTTTAGACAGCATCAAACATTCAGAACTGCTATTGGATATATCGGAGTGTGATAAGGACGATATCACACGATACGCGGACCGCATCCTGTCCCGGGCTATGACCGTAGAGGTGACAGTGCGCACTGACCGAGACCATCAGCAGGAGGAAGCTCTGTATCAGGTGAACATGTACATCGATCAGCTGGTGATGTCCGTGCACAACGACGCAGTGAGTGCGCATTCCCGTTGCCAGACCTATATGAACGCGTGCACGTCTCAGCCGGACCCCAACGCGGGCACCGACAAGAACTTTGAGACGGCCATCCTGGGCTGCACCCTCGACGATCAGAAGCGCGTCAAGAAACGACTCCAAGGCCTGCTGGACTACATCGCGAAGTTAAACGTGACCACGTGCTCGTGA
Protein
MRLIAVLDQVEMRVERLRKDTVRIEEEKDSLLSTLDSIKHSELLLDISECDKDDITRYADRILSRAMTVEVTVRTDRDHQQEEALYQVNMYIDQLVMSVHNDAVSAHSRCQTYMNACTSQPDPNAGTDKNFETAILGCTLDDQKRVKKRLQGLLDYIAKLNVTTCS
Summary
Uniprot
H9JX14
A0A194QVL1
A0A194QES2
A0A2H1VHS7
A0A2W1BT16
A0A2A4J591
+ More
A0A3S2LPY5 S4P9Y9 A0A212F6V1 A0A2R7WQF1 A0A1W4XB55 A0A1W4XKP1 E2B7Q8 A0A0V0G8A5 A0A224XQC4 A0A069DNZ0 D6WT70 A0A023F9S6 A0A0P4VT04 R4FLC6 A0A195CB31 A0A2J7Q8M2 A0A026WX79 A0A151X6T7 A0A195FEN6 A0A0A9Z481 A0A0A9Z6I9 A0A0A9ZE26 F4WY87 A0A2P8XQY3 A0A195EHF8 A0A195AWJ8 A0A158NHR7 A0A1B6EFD8 E0VQI7 A0A0K8SGE9 A0A0A9Z474 A0A1Y1LPQ5 A0A087ZQI4 A0A1Y1LPU6 V5IA55 K7J5H0 A0A232EWC6 A0A2A3E4S0 V9IKN3 A0A1B6M1T6 A0A067QRB2 J3JUE1 N6U2I9 A0A1B6JKT9 A0A0J7NZD5 A0A0L7QXV9 A0A0T6BBR6 A0A154P903 A0A0M9A912 U4ULN7 E2AZR5 A0A0Q9XQ06 A0A0Q9XCR6 A0A0Q9WW09 B4KUI7 B4LH78 A0A1W4VBE2 A0A0Q5UJN2 A0A1W4VNT9 B4NLW2 A0A0P8XVT9 A0A0R1DYH1 A0A3B0K8T1 A0A0R1EDL2 B4IU08 B4PCG6 B3NI32 B3M703 A0A0Q5U5Z6 Q176J7 A0A3B0KAK5 Q176J8 Q9VUQ1 Q176J6 A0A3B0KWL2 B4QL28 Q95RY2 A0A0Q9WJC7 A0A0L7LJJ3 B4IWV8 A0A0M4F1B9 A0A0J9RWX3 A0A3B0K6K6 A0A1B0D952 A0A1L8DTR9 A0A1S3CVN0 A0A0R3P7F5 Q2M1C5 A0A0L0C8F3 U5EPL7 A0A1Q3FAU1 A0A0K8WHG3
A0A3S2LPY5 S4P9Y9 A0A212F6V1 A0A2R7WQF1 A0A1W4XB55 A0A1W4XKP1 E2B7Q8 A0A0V0G8A5 A0A224XQC4 A0A069DNZ0 D6WT70 A0A023F9S6 A0A0P4VT04 R4FLC6 A0A195CB31 A0A2J7Q8M2 A0A026WX79 A0A151X6T7 A0A195FEN6 A0A0A9Z481 A0A0A9Z6I9 A0A0A9ZE26 F4WY87 A0A2P8XQY3 A0A195EHF8 A0A195AWJ8 A0A158NHR7 A0A1B6EFD8 E0VQI7 A0A0K8SGE9 A0A0A9Z474 A0A1Y1LPQ5 A0A087ZQI4 A0A1Y1LPU6 V5IA55 K7J5H0 A0A232EWC6 A0A2A3E4S0 V9IKN3 A0A1B6M1T6 A0A067QRB2 J3JUE1 N6U2I9 A0A1B6JKT9 A0A0J7NZD5 A0A0L7QXV9 A0A0T6BBR6 A0A154P903 A0A0M9A912 U4ULN7 E2AZR5 A0A0Q9XQ06 A0A0Q9XCR6 A0A0Q9WW09 B4KUI7 B4LH78 A0A1W4VBE2 A0A0Q5UJN2 A0A1W4VNT9 B4NLW2 A0A0P8XVT9 A0A0R1DYH1 A0A3B0K8T1 A0A0R1EDL2 B4IU08 B4PCG6 B3NI32 B3M703 A0A0Q5U5Z6 Q176J7 A0A3B0KAK5 Q176J8 Q9VUQ1 Q176J6 A0A3B0KWL2 B4QL28 Q95RY2 A0A0Q9WJC7 A0A0L7LJJ3 B4IWV8 A0A0M4F1B9 A0A0J9RWX3 A0A3B0K6K6 A0A1B0D952 A0A1L8DTR9 A0A1S3CVN0 A0A0R3P7F5 Q2M1C5 A0A0L0C8F3 U5EPL7 A0A1Q3FAU1 A0A0K8WHG3
Pubmed
19121390
26354079
28756777
23622113
22118469
20798317
+ More
26334808 18362917 19820115 25474469 27129103 24508170 25401762 26823975 21719571 29403074 21347285 20566863 28004739 20075255 28648823 24845553 22516182 23537049 17994087 18057021 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26227816 15632085 23185243 26108605
26334808 18362917 19820115 25474469 27129103 24508170 25401762 26823975 21719571 29403074 21347285 20566863 28004739 20075255 28648823 24845553 22516182 23537049 17994087 18057021 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26227816 15632085 23185243 26108605
EMBL
BABH01042529
BABH01042530
KQ461073
KPJ09573.1
KQ459053
KPJ04038.1
+ More
ODYU01002616 SOQ40346.1 KZ149982 PZC75776.1 NWSH01002914 PCG67307.1 RSAL01000034 RVE51416.1 GAIX01003494 JAA89066.1 AGBW02009975 OWR49439.1 KK855279 PTY21797.1 GL446201 EFN88271.1 GECL01001745 JAP04379.1 GFTR01003142 JAW13284.1 GBGD01003101 JAC85788.1 KQ971354 EFA05854.1 GBBI01001034 JAC17678.1 GDKW01002510 JAI54085.1 ACPB03013538 GAHY01002154 JAA75356.1 KQ978023 KYM98069.1 NEVH01016952 PNF24923.1 KK107078 EZA60448.1 KQ982476 KYQ56072.1 KQ981636 KYN38863.1 GBHO01003533 GBHO01003532 JAG40071.1 JAG40072.1 GBHO01003540 GBHO01003536 GBHO01003535 GBHO01003424 GBRD01013459 GDHC01020044 JAG40064.1 JAG40068.1 JAG40069.1 JAG40180.1 JAG52367.1 JAP98584.1 GBHO01003539 GBHO01003534 GBRD01013457 JAG40065.1 JAG40070.1 JAG52369.1 GL888439 EGI60844.1 PYGN01001502 PSN34410.1 KQ978957 KYN27294.1 KQ976725 KYM76623.1 ADTU01016033 GEDC01000632 JAS36666.1 DS235430 EEB15643.1 GBRD01013458 JAG52368.1 GBHO01003537 GBHO01003531 JAG40067.1 JAG40073.1 GEZM01050234 JAV75664.1 GEZM01050235 JAV75663.1 GALX01001775 JAB66691.1 NNAY01001882 OXU22654.1 KZ288386 PBC26484.1 JR052422 AEY61693.1 GEBQ01010092 JAT29885.1 KK853035 KDR12234.1 BT126854 AEE61816.1 APGK01052222 APGK01052223 KB741211 ENN72777.1 GECU01020056 GECU01015868 GECU01009173 GECU01007815 JAS87650.1 JAS91838.1 JAS98533.1 JAS99891.1 LBMM01000686 KMQ97760.1 KQ414699 KOC63453.1 LJIG01002148 KRT84792.1 KQ434846 KZC08323.1 KQ435724 KOX78349.1 KB632263 ERL90910.1 GL444277 EFN61044.1 CH933809 KRG06067.1 KRG06069.1 CH940647 KRF85003.1 EDW18215.1 KRG06068.1 EDW70591.2 KRF85002.1 CH954178 KQS44106.1 CH964278 EDW85414.1 CH902618 KPU78822.1 CM000159 KRK02093.1 OUUW01000027 SPP89753.1 CH891752 KRK05268.1 EDW99871.2 EDW94876.2 EDV52118.2 EDV40868.2 KQS44105.1 CH477386 EAT42086.1 SPP89752.1 EAT42087.1 AE014296 BT011355 AAF49625.1 AAR96147.1 EAT42084.1 EAT42085.1 SPP89751.1 CM000363 CM002912 EDX10553.1 KMY99758.1 AY061044 AAF49626.2 AAL28592.1 KRF85004.1 JTDY01000937 KOB75366.1 CH916366 EDV97359.1 CP012525 ALC44707.1 KMY99759.1 SPP89754.1 AJVK01012911 GFDF01004225 JAV09859.1 CH379069 KRT07501.1 EAL30649.3 KRT07502.1 JRES01000760 KNC28507.1 GANO01003670 JAB56201.1 GFDL01010432 JAV24613.1 GDHF01029625 GDHF01017154 GDHF01006254 GDHF01001782 JAI22689.1 JAI35160.1 JAI46060.1 JAI50532.1
ODYU01002616 SOQ40346.1 KZ149982 PZC75776.1 NWSH01002914 PCG67307.1 RSAL01000034 RVE51416.1 GAIX01003494 JAA89066.1 AGBW02009975 OWR49439.1 KK855279 PTY21797.1 GL446201 EFN88271.1 GECL01001745 JAP04379.1 GFTR01003142 JAW13284.1 GBGD01003101 JAC85788.1 KQ971354 EFA05854.1 GBBI01001034 JAC17678.1 GDKW01002510 JAI54085.1 ACPB03013538 GAHY01002154 JAA75356.1 KQ978023 KYM98069.1 NEVH01016952 PNF24923.1 KK107078 EZA60448.1 KQ982476 KYQ56072.1 KQ981636 KYN38863.1 GBHO01003533 GBHO01003532 JAG40071.1 JAG40072.1 GBHO01003540 GBHO01003536 GBHO01003535 GBHO01003424 GBRD01013459 GDHC01020044 JAG40064.1 JAG40068.1 JAG40069.1 JAG40180.1 JAG52367.1 JAP98584.1 GBHO01003539 GBHO01003534 GBRD01013457 JAG40065.1 JAG40070.1 JAG52369.1 GL888439 EGI60844.1 PYGN01001502 PSN34410.1 KQ978957 KYN27294.1 KQ976725 KYM76623.1 ADTU01016033 GEDC01000632 JAS36666.1 DS235430 EEB15643.1 GBRD01013458 JAG52368.1 GBHO01003537 GBHO01003531 JAG40067.1 JAG40073.1 GEZM01050234 JAV75664.1 GEZM01050235 JAV75663.1 GALX01001775 JAB66691.1 NNAY01001882 OXU22654.1 KZ288386 PBC26484.1 JR052422 AEY61693.1 GEBQ01010092 JAT29885.1 KK853035 KDR12234.1 BT126854 AEE61816.1 APGK01052222 APGK01052223 KB741211 ENN72777.1 GECU01020056 GECU01015868 GECU01009173 GECU01007815 JAS87650.1 JAS91838.1 JAS98533.1 JAS99891.1 LBMM01000686 KMQ97760.1 KQ414699 KOC63453.1 LJIG01002148 KRT84792.1 KQ434846 KZC08323.1 KQ435724 KOX78349.1 KB632263 ERL90910.1 GL444277 EFN61044.1 CH933809 KRG06067.1 KRG06069.1 CH940647 KRF85003.1 EDW18215.1 KRG06068.1 EDW70591.2 KRF85002.1 CH954178 KQS44106.1 CH964278 EDW85414.1 CH902618 KPU78822.1 CM000159 KRK02093.1 OUUW01000027 SPP89753.1 CH891752 KRK05268.1 EDW99871.2 EDW94876.2 EDV52118.2 EDV40868.2 KQS44105.1 CH477386 EAT42086.1 SPP89752.1 EAT42087.1 AE014296 BT011355 AAF49625.1 AAR96147.1 EAT42084.1 EAT42085.1 SPP89751.1 CM000363 CM002912 EDX10553.1 KMY99758.1 AY061044 AAF49626.2 AAL28592.1 KRF85004.1 JTDY01000937 KOB75366.1 CH916366 EDV97359.1 CP012525 ALC44707.1 KMY99759.1 SPP89754.1 AJVK01012911 GFDF01004225 JAV09859.1 CH379069 KRT07501.1 EAL30649.3 KRT07502.1 JRES01000760 KNC28507.1 GANO01003670 JAB56201.1 GFDL01010432 JAV24613.1 GDHF01029625 GDHF01017154 GDHF01006254 GDHF01001782 JAI22689.1 JAI35160.1 JAI46060.1 JAI50532.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000192223 UP000008237 UP000007266 UP000015103 UP000078542 UP000235965 UP000053097 UP000075809 UP000078541 UP000007755 UP000245037 UP000078492 UP000078540 UP000005205 UP000009046 UP000005203 UP000002358 UP000215335 UP000242457 UP000027135 UP000019118 UP000036403 UP000053825 UP000076502 UP000053105 UP000030742 UP000000311 UP000009192 UP000008792 UP000192221 UP000008711 UP000007798 UP000007801 UP000002282 UP000268350 UP000008820 UP000000803 UP000000304 UP000037510 UP000001070 UP000092553 UP000092462 UP000079169 UP000001819 UP000037069
UP000192223 UP000008237 UP000007266 UP000015103 UP000078542 UP000235965 UP000053097 UP000075809 UP000078541 UP000007755 UP000245037 UP000078492 UP000078540 UP000005205 UP000009046 UP000005203 UP000002358 UP000215335 UP000242457 UP000027135 UP000019118 UP000036403 UP000053825 UP000076502 UP000053105 UP000030742 UP000000311 UP000009192 UP000008792 UP000192221 UP000008711 UP000007798 UP000007801 UP000002282 UP000268350 UP000008820 UP000000803 UP000000304 UP000037510 UP000001070 UP000092553 UP000092462 UP000079169 UP000001819 UP000037069
PRIDE
Interpro
ProteinModelPortal
H9JX14
A0A194QVL1
A0A194QES2
A0A2H1VHS7
A0A2W1BT16
A0A2A4J591
+ More
A0A3S2LPY5 S4P9Y9 A0A212F6V1 A0A2R7WQF1 A0A1W4XB55 A0A1W4XKP1 E2B7Q8 A0A0V0G8A5 A0A224XQC4 A0A069DNZ0 D6WT70 A0A023F9S6 A0A0P4VT04 R4FLC6 A0A195CB31 A0A2J7Q8M2 A0A026WX79 A0A151X6T7 A0A195FEN6 A0A0A9Z481 A0A0A9Z6I9 A0A0A9ZE26 F4WY87 A0A2P8XQY3 A0A195EHF8 A0A195AWJ8 A0A158NHR7 A0A1B6EFD8 E0VQI7 A0A0K8SGE9 A0A0A9Z474 A0A1Y1LPQ5 A0A087ZQI4 A0A1Y1LPU6 V5IA55 K7J5H0 A0A232EWC6 A0A2A3E4S0 V9IKN3 A0A1B6M1T6 A0A067QRB2 J3JUE1 N6U2I9 A0A1B6JKT9 A0A0J7NZD5 A0A0L7QXV9 A0A0T6BBR6 A0A154P903 A0A0M9A912 U4ULN7 E2AZR5 A0A0Q9XQ06 A0A0Q9XCR6 A0A0Q9WW09 B4KUI7 B4LH78 A0A1W4VBE2 A0A0Q5UJN2 A0A1W4VNT9 B4NLW2 A0A0P8XVT9 A0A0R1DYH1 A0A3B0K8T1 A0A0R1EDL2 B4IU08 B4PCG6 B3NI32 B3M703 A0A0Q5U5Z6 Q176J7 A0A3B0KAK5 Q176J8 Q9VUQ1 Q176J6 A0A3B0KWL2 B4QL28 Q95RY2 A0A0Q9WJC7 A0A0L7LJJ3 B4IWV8 A0A0M4F1B9 A0A0J9RWX3 A0A3B0K6K6 A0A1B0D952 A0A1L8DTR9 A0A1S3CVN0 A0A0R3P7F5 Q2M1C5 A0A0L0C8F3 U5EPL7 A0A1Q3FAU1 A0A0K8WHG3
A0A3S2LPY5 S4P9Y9 A0A212F6V1 A0A2R7WQF1 A0A1W4XB55 A0A1W4XKP1 E2B7Q8 A0A0V0G8A5 A0A224XQC4 A0A069DNZ0 D6WT70 A0A023F9S6 A0A0P4VT04 R4FLC6 A0A195CB31 A0A2J7Q8M2 A0A026WX79 A0A151X6T7 A0A195FEN6 A0A0A9Z481 A0A0A9Z6I9 A0A0A9ZE26 F4WY87 A0A2P8XQY3 A0A195EHF8 A0A195AWJ8 A0A158NHR7 A0A1B6EFD8 E0VQI7 A0A0K8SGE9 A0A0A9Z474 A0A1Y1LPQ5 A0A087ZQI4 A0A1Y1LPU6 V5IA55 K7J5H0 A0A232EWC6 A0A2A3E4S0 V9IKN3 A0A1B6M1T6 A0A067QRB2 J3JUE1 N6U2I9 A0A1B6JKT9 A0A0J7NZD5 A0A0L7QXV9 A0A0T6BBR6 A0A154P903 A0A0M9A912 U4ULN7 E2AZR5 A0A0Q9XQ06 A0A0Q9XCR6 A0A0Q9WW09 B4KUI7 B4LH78 A0A1W4VBE2 A0A0Q5UJN2 A0A1W4VNT9 B4NLW2 A0A0P8XVT9 A0A0R1DYH1 A0A3B0K8T1 A0A0R1EDL2 B4IU08 B4PCG6 B3NI32 B3M703 A0A0Q5U5Z6 Q176J7 A0A3B0KAK5 Q176J8 Q9VUQ1 Q176J6 A0A3B0KWL2 B4QL28 Q95RY2 A0A0Q9WJC7 A0A0L7LJJ3 B4IWV8 A0A0M4F1B9 A0A0J9RWX3 A0A3B0K6K6 A0A1B0D952 A0A1L8DTR9 A0A1S3CVN0 A0A0R3P7F5 Q2M1C5 A0A0L0C8F3 U5EPL7 A0A1Q3FAU1 A0A0K8WHG3
PDB
3D0T
E-value=9.11427e-07,
Score=121
Ontologies
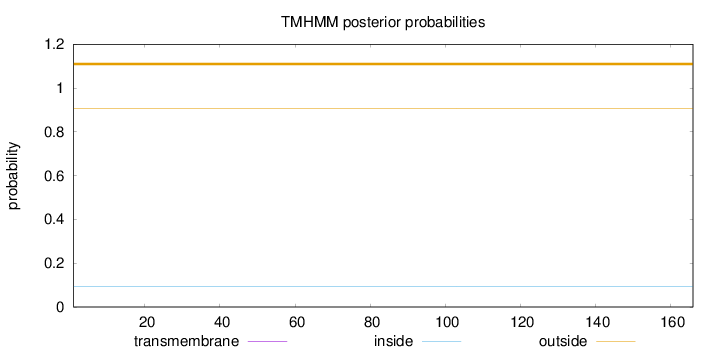
Topology
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09396
outside
1 - 166
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
