Gene
KWMTBOMO16670
Annotation
PREDICTED:_uncharacterized_protein_LOC100101190_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.948
Sequence
CDS
ATGTTTGTGGTATCTCCTCATATACGCCAGGAATGGAAATGTATGGCTACTCTGATTGCAAGATTTTCAAATAAACCCACATCATCCGAGGTTCTAACAGCCTATTTAACTCAGTGTAACGAACCACCTTGGACTTCTTATTTTGTAAAGTACAGTTATGTGAAAGATGATCAGTTTGGAATGTCAAATTTTAATTGGAAGGTTGGTAATTCCAACTATCAAATATTAAGAACTGGTTGTTTTCCATATATAAAATATCACTGTTCTAGAAAAAAGGCAGAAGACTTAAATATGTCAGATAAATTTATGAGAATTATAAAGGTTGCAAATTTAGGTATACCTTGTTTACTGTATGGCCTTGCAGCAACCCAGCTTATAAGGCATGAAGAACTTGTTCACACAAGCAAAGGGCCAGTACCAATATACTTTTTGCTTCCAGAAGATAAAGGATCTTTGCATTAA
Protein
MFVVSPHIRQEWKCMATLIARFSNKPTSSEVLTAYLTQCNEPPWTSYFVKYSYVKDDQFGMSNFNWKVGNSNYQILRTGCFPYIKYHCSRKKAEDLNMSDKFMRIIKVANLGIPCLLYGLAATQLIRHEELVHTSKGPVPIYFLLPEDKGSLH
Summary
Similarity
Belongs to the complex I 49 kDa subunit family.
Uniprot
A0A2A4JTL9
A0A2H1VJQ0
A0A0L7LI69
A0A212ESJ4
A0A194RN77
A0A194QBK8
+ More
A0A3S2LRI7 A0A1Y1LB50 A0A0C9S0V9 A0A026WFQ6 A0A151WUY8 A0A232FL96 A0A067QMQ1 A0A195EXY9 A0A2P8YI59 K7IY11 A0A158NMT3 A0A195BC03 A0A139WF30 A0A088AFK1 A0A151JM52 F4WTC2 A0A2J7R6I9 A0A1B6D519 A0A1W4WY99 E2C9N7 A0A0K8UAY1 A0NCR8 A0A182X5B4 A0A182TN49 A0A182I9M1 A0A182V7W2 E2ALJ3 A0A154P4F0 A0A0A9WY25 A0A182PM66 A0A182KFB2 A0A1D2NC66 A4V381 B3DNH9 A0A2M4AZQ4 A0A182J5B4 A0A195C7N9 A0A2H8TNN0 A0A182SRP4 J9JRW7 A0A182XY40 A0A293MFV5 A0A182QJI2 B0WWT0 A0A182FIL7 V9L4Z7 A0A182N0Y9 A0A1I8N5H7 W5JHJ8 A0A182MPF4 T1DND8 A0A084WPL8 A7RSK0 A0A1A9WY09 A0A1I8NYZ6 A0A1B0BYC9 A0A0K8TLP7 E9GMQ1 A0A3B5KE75 A0A182RJ19 A0A226E833 A0A3Q3JA43 A0A0P5GUT6 A0A1A9YLP5 A0A1S4FTD6 A0A182LD63 Q16Q67 A0A3P9GYR6 A0A182VVB4 A0A3B5KHR3 A0A3P9KFJ5 A0A224Y4C4 A0A2G9RCA1 A0A3B3DWC1 A0A3Q3VR30 U5EH96 H3AWY9 A0A3S2Q6P1 Q4SLW5 A0A3B4YLD5 H2LQW6 A0A131Y3M4 A0A0F8BV48 A0A3B4TYB8 A0A3Q3L5E8 A0A1A7WGC2 B7P7S6 A0A3B4GCR5 A0A3Q2W3W9 A0A3Q4MV60 A0A3P9D7C7 A0A3P8U3H4 A0A3Q3DH82
A0A3S2LRI7 A0A1Y1LB50 A0A0C9S0V9 A0A026WFQ6 A0A151WUY8 A0A232FL96 A0A067QMQ1 A0A195EXY9 A0A2P8YI59 K7IY11 A0A158NMT3 A0A195BC03 A0A139WF30 A0A088AFK1 A0A151JM52 F4WTC2 A0A2J7R6I9 A0A1B6D519 A0A1W4WY99 E2C9N7 A0A0K8UAY1 A0NCR8 A0A182X5B4 A0A182TN49 A0A182I9M1 A0A182V7W2 E2ALJ3 A0A154P4F0 A0A0A9WY25 A0A182PM66 A0A182KFB2 A0A1D2NC66 A4V381 B3DNH9 A0A2M4AZQ4 A0A182J5B4 A0A195C7N9 A0A2H8TNN0 A0A182SRP4 J9JRW7 A0A182XY40 A0A293MFV5 A0A182QJI2 B0WWT0 A0A182FIL7 V9L4Z7 A0A182N0Y9 A0A1I8N5H7 W5JHJ8 A0A182MPF4 T1DND8 A0A084WPL8 A7RSK0 A0A1A9WY09 A0A1I8NYZ6 A0A1B0BYC9 A0A0K8TLP7 E9GMQ1 A0A3B5KE75 A0A182RJ19 A0A226E833 A0A3Q3JA43 A0A0P5GUT6 A0A1A9YLP5 A0A1S4FTD6 A0A182LD63 Q16Q67 A0A3P9GYR6 A0A182VVB4 A0A3B5KHR3 A0A3P9KFJ5 A0A224Y4C4 A0A2G9RCA1 A0A3B3DWC1 A0A3Q3VR30 U5EH96 H3AWY9 A0A3S2Q6P1 Q4SLW5 A0A3B4YLD5 H2LQW6 A0A131Y3M4 A0A0F8BV48 A0A3B4TYB8 A0A3Q3L5E8 A0A1A7WGC2 B7P7S6 A0A3B4GCR5 A0A3Q2W3W9 A0A3Q4MV60 A0A3P9D7C7 A0A3P8U3H4 A0A3Q3DH82
Pubmed
26227816
22118469
26354079
28004739
24508170
30249741
+ More
28648823 24845553 29403074 20075255 21347285 18362917 19820115 21719571 20798317 12364791 14747013 17210077 25401762 27289101 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 24402279 25315136 20920257 23761445 24438588 17615350 26369729 21292972 21551351 20966253 17510324 17554307 28797301 29451363 9215903 15496914 25835551 25186727
28648823 24845553 29403074 20075255 21347285 18362917 19820115 21719571 20798317 12364791 14747013 17210077 25401762 27289101 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 24402279 25315136 20920257 23761445 24438588 17615350 26369729 21292972 21551351 20966253 17510324 17554307 28797301 29451363 9215903 15496914 25835551 25186727
EMBL
NWSH01000683
PCG74813.1
ODYU01002932
SOQ41048.1
JTDY01001004
KOB75142.1
+ More
AGBW02012778 OWR44460.1 KQ459984 KPJ18967.1 KQ459220 KPJ02847.1 RSAL01000293 RVE42876.1 GEZM01062046 JAV70091.1 GBYB01014251 JAG84018.1 KK107245 QOIP01000014 EZA54506.1 RLU14821.1 KQ982729 KYQ51577.1 NNAY01000071 OXU31279.1 KK853153 KDR10553.1 KQ981940 KYN32744.1 PYGN01000580 PSN43884.1 ADTU01020800 KQ976529 KYM81725.1 KQ971354 KYB26580.1 KQ978946 KYN27401.1 GL888334 EGI62585.1 NEVH01006756 PNF36448.1 GEDC01016555 GEDC01013341 JAS20743.1 JAS23957.1 GL453868 EFN75376.1 GDHF01028799 JAI23515.1 AAAB01008847 EAU77223.3 APCN01001005 GL440609 EFN65695.1 KQ434810 KZC06737.1 GBHO01031273 GBHO01011758 GBRD01004528 JAG12331.1 JAG31846.1 JAG61293.1 LJIJ01000099 ODN02685.1 AE014297 AAN13890.2 BT032967 ACD99531.1 GGFK01012948 MBW46269.1 KQ978231 KYM96163.1 GFXV01003063 MBW14868.1 ABLF02019454 GFWV01019829 MAA44557.1 AXCN02001373 DS232152 EDS36191.1 JW874163 AFP06680.1 ADMH02001172 ETN63842.1 AXCM01009391 GAMD01002975 JAA98615.1 ATLV01025101 KE525369 KFB52162.1 DS469534 EDO45650.1 JXJN01022584 GDAI01002339 JAI15264.1 GL732553 EFX79255.1 LNIX01000006 OXA52816.1 GDIQ01258686 JAJ93038.1 CH477759 EAT36539.1 GFPF01001272 MAA12418.1 KV946802 PIO25516.1 GANO01003121 JAB56750.1 AFYH01123882 CM012442 RVE71806.1 CAAE01014555 CAF98367.1 GEFM01001947 JAP73849.1 KQ042192 KKF18153.1 HADW01003211 HADX01006543 SBP04611.1 ABJB010944827 DS653188 EEC02648.1
AGBW02012778 OWR44460.1 KQ459984 KPJ18967.1 KQ459220 KPJ02847.1 RSAL01000293 RVE42876.1 GEZM01062046 JAV70091.1 GBYB01014251 JAG84018.1 KK107245 QOIP01000014 EZA54506.1 RLU14821.1 KQ982729 KYQ51577.1 NNAY01000071 OXU31279.1 KK853153 KDR10553.1 KQ981940 KYN32744.1 PYGN01000580 PSN43884.1 ADTU01020800 KQ976529 KYM81725.1 KQ971354 KYB26580.1 KQ978946 KYN27401.1 GL888334 EGI62585.1 NEVH01006756 PNF36448.1 GEDC01016555 GEDC01013341 JAS20743.1 JAS23957.1 GL453868 EFN75376.1 GDHF01028799 JAI23515.1 AAAB01008847 EAU77223.3 APCN01001005 GL440609 EFN65695.1 KQ434810 KZC06737.1 GBHO01031273 GBHO01011758 GBRD01004528 JAG12331.1 JAG31846.1 JAG61293.1 LJIJ01000099 ODN02685.1 AE014297 AAN13890.2 BT032967 ACD99531.1 GGFK01012948 MBW46269.1 KQ978231 KYM96163.1 GFXV01003063 MBW14868.1 ABLF02019454 GFWV01019829 MAA44557.1 AXCN02001373 DS232152 EDS36191.1 JW874163 AFP06680.1 ADMH02001172 ETN63842.1 AXCM01009391 GAMD01002975 JAA98615.1 ATLV01025101 KE525369 KFB52162.1 DS469534 EDO45650.1 JXJN01022584 GDAI01002339 JAI15264.1 GL732553 EFX79255.1 LNIX01000006 OXA52816.1 GDIQ01258686 JAJ93038.1 CH477759 EAT36539.1 GFPF01001272 MAA12418.1 KV946802 PIO25516.1 GANO01003121 JAB56750.1 AFYH01123882 CM012442 RVE71806.1 CAAE01014555 CAF98367.1 GEFM01001947 JAP73849.1 KQ042192 KKF18153.1 HADW01003211 HADX01006543 SBP04611.1 ABJB010944827 DS653188 EEC02648.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000053097 UP000279307 UP000075809 UP000215335 UP000027135 UP000078541 UP000245037 UP000002358 UP000005205 UP000078540 UP000007266 UP000005203 UP000078492 UP000007755 UP000235965 UP000192223 UP000008237 UP000007062 UP000076407 UP000075902 UP000075840 UP000075903 UP000000311 UP000076502 UP000075885 UP000075881 UP000094527 UP000000803 UP000075880 UP000078542 UP000075901 UP000007819 UP000076408 UP000075886 UP000002320 UP000069272 UP000075884 UP000095301 UP000000673 UP000075883 UP000030765 UP000001593 UP000091820 UP000095300 UP000092460 UP000000305 UP000005226 UP000075900 UP000198287 UP000261600 UP000092443 UP000075882 UP000008820 UP000265200 UP000075920 UP000265180 UP000261560 UP000261620 UP000008672 UP000261360 UP000001038 UP000261420 UP000261640 UP000001555 UP000261460 UP000264840 UP000261580 UP000265160 UP000265080 UP000264820
UP000053097 UP000279307 UP000075809 UP000215335 UP000027135 UP000078541 UP000245037 UP000002358 UP000005205 UP000078540 UP000007266 UP000005203 UP000078492 UP000007755 UP000235965 UP000192223 UP000008237 UP000007062 UP000076407 UP000075902 UP000075840 UP000075903 UP000000311 UP000076502 UP000075885 UP000075881 UP000094527 UP000000803 UP000075880 UP000078542 UP000075901 UP000007819 UP000076408 UP000075886 UP000002320 UP000069272 UP000075884 UP000095301 UP000000673 UP000075883 UP000030765 UP000001593 UP000091820 UP000095300 UP000092460 UP000000305 UP000005226 UP000075900 UP000198287 UP000261600 UP000092443 UP000075882 UP000008820 UP000265200 UP000075920 UP000265180 UP000261560 UP000261620 UP000008672 UP000261360 UP000001038 UP000261420 UP000261640 UP000001555 UP000261460 UP000264840 UP000261580 UP000265160 UP000265080 UP000264820
PRIDE
Interpro
SUPFAM
SSF56762
SSF56762
Gene 3D
ProteinModelPortal
A0A2A4JTL9
A0A2H1VJQ0
A0A0L7LI69
A0A212ESJ4
A0A194RN77
A0A194QBK8
+ More
A0A3S2LRI7 A0A1Y1LB50 A0A0C9S0V9 A0A026WFQ6 A0A151WUY8 A0A232FL96 A0A067QMQ1 A0A195EXY9 A0A2P8YI59 K7IY11 A0A158NMT3 A0A195BC03 A0A139WF30 A0A088AFK1 A0A151JM52 F4WTC2 A0A2J7R6I9 A0A1B6D519 A0A1W4WY99 E2C9N7 A0A0K8UAY1 A0NCR8 A0A182X5B4 A0A182TN49 A0A182I9M1 A0A182V7W2 E2ALJ3 A0A154P4F0 A0A0A9WY25 A0A182PM66 A0A182KFB2 A0A1D2NC66 A4V381 B3DNH9 A0A2M4AZQ4 A0A182J5B4 A0A195C7N9 A0A2H8TNN0 A0A182SRP4 J9JRW7 A0A182XY40 A0A293MFV5 A0A182QJI2 B0WWT0 A0A182FIL7 V9L4Z7 A0A182N0Y9 A0A1I8N5H7 W5JHJ8 A0A182MPF4 T1DND8 A0A084WPL8 A7RSK0 A0A1A9WY09 A0A1I8NYZ6 A0A1B0BYC9 A0A0K8TLP7 E9GMQ1 A0A3B5KE75 A0A182RJ19 A0A226E833 A0A3Q3JA43 A0A0P5GUT6 A0A1A9YLP5 A0A1S4FTD6 A0A182LD63 Q16Q67 A0A3P9GYR6 A0A182VVB4 A0A3B5KHR3 A0A3P9KFJ5 A0A224Y4C4 A0A2G9RCA1 A0A3B3DWC1 A0A3Q3VR30 U5EH96 H3AWY9 A0A3S2Q6P1 Q4SLW5 A0A3B4YLD5 H2LQW6 A0A131Y3M4 A0A0F8BV48 A0A3B4TYB8 A0A3Q3L5E8 A0A1A7WGC2 B7P7S6 A0A3B4GCR5 A0A3Q2W3W9 A0A3Q4MV60 A0A3P9D7C7 A0A3P8U3H4 A0A3Q3DH82
A0A3S2LRI7 A0A1Y1LB50 A0A0C9S0V9 A0A026WFQ6 A0A151WUY8 A0A232FL96 A0A067QMQ1 A0A195EXY9 A0A2P8YI59 K7IY11 A0A158NMT3 A0A195BC03 A0A139WF30 A0A088AFK1 A0A151JM52 F4WTC2 A0A2J7R6I9 A0A1B6D519 A0A1W4WY99 E2C9N7 A0A0K8UAY1 A0NCR8 A0A182X5B4 A0A182TN49 A0A182I9M1 A0A182V7W2 E2ALJ3 A0A154P4F0 A0A0A9WY25 A0A182PM66 A0A182KFB2 A0A1D2NC66 A4V381 B3DNH9 A0A2M4AZQ4 A0A182J5B4 A0A195C7N9 A0A2H8TNN0 A0A182SRP4 J9JRW7 A0A182XY40 A0A293MFV5 A0A182QJI2 B0WWT0 A0A182FIL7 V9L4Z7 A0A182N0Y9 A0A1I8N5H7 W5JHJ8 A0A182MPF4 T1DND8 A0A084WPL8 A7RSK0 A0A1A9WY09 A0A1I8NYZ6 A0A1B0BYC9 A0A0K8TLP7 E9GMQ1 A0A3B5KE75 A0A182RJ19 A0A226E833 A0A3Q3JA43 A0A0P5GUT6 A0A1A9YLP5 A0A1S4FTD6 A0A182LD63 Q16Q67 A0A3P9GYR6 A0A182VVB4 A0A3B5KHR3 A0A3P9KFJ5 A0A224Y4C4 A0A2G9RCA1 A0A3B3DWC1 A0A3Q3VR30 U5EH96 H3AWY9 A0A3S2Q6P1 Q4SLW5 A0A3B4YLD5 H2LQW6 A0A131Y3M4 A0A0F8BV48 A0A3B4TYB8 A0A3Q3L5E8 A0A1A7WGC2 B7P7S6 A0A3B4GCR5 A0A3Q2W3W9 A0A3Q4MV60 A0A3P9D7C7 A0A3P8U3H4 A0A3Q3DH82
Ontologies
PANTHER
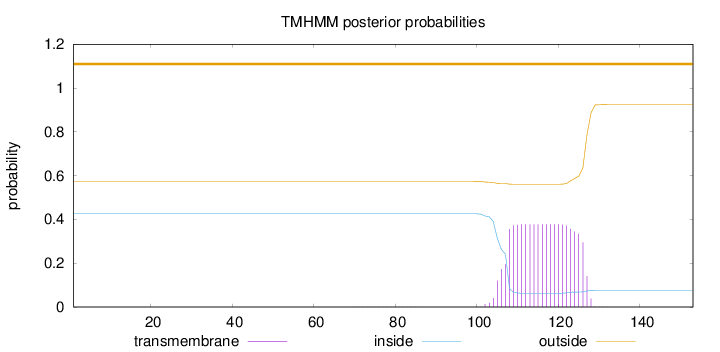
Topology
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.71244
Exp number, first 60 AAs:
0.00145
Total prob of N-in:
0.42573
outside
1 - 153
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
