Gene
KWMTBOMO16669 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013349
Annotation
PREDICTED:_adapter_molecule_Crk_isoform_X1_[Papilio_polytes]
Full name
Adapter molecule Crk
Location in the cell
Cytoplasmic Reliability : 1.122
Sequence
CDS
ATGGCGAATTCCGGAATGGGGGGTTCCTTCGACCCCAATGACATGAGCAGCTGGTACTTTCACGGCATGACGCGGAAAGAAGCTACAGATCTGCTCCTTAATGAGACTGAGAGTGGCGTGTTTCTGGTCAGGGATTCGAAGTCCATTGTTGGAAACTACGTTCTTTGCGTCAAAGAAGCAGGTCGGGTGTCGAACTATATAATCGATCGCATCGTTTCACCCGACGGCTCGGTGCTCTTCCGCATCGGAGACCAGGTGTTCGTGGACATGGCGGCGCTGTTGTCGTTCTACCGCGTCCACTACTTGGACACGACGGCGCTGGTGAGGCCCTACGCGAGGACGGCCATCGCTCACCAGCCCTCCGCAGCAGAACCAGACCCTAGAGATAGTTATCGCGAAGTTCGATTTCGCTGGCAACGTGAGTAA
Protein
MANSGMGGSFDPNDMSSWYFHGMTRKEATDLLLNETESGVFLVRDSKSIVGNYVLCVKEAGRVSNYIIDRIVSPDGSVLFRIGDQVFVDMAALLSFYRVHYLDTTALVRPYARTAIAHQPSAAEPDPRDSYREVRFRWQRE
Summary
Description
Adapter protein which interacts with C-terminal portion of mbc, homolog of human DOCK180. May play a role in cellular processes throughout development.
Similarity
Belongs to the CRK family.
Keywords
Complete proteome
Developmental protein
Reference proteome
Repeat
SH2 domain
SH3 domain
Feature
chain Adapter molecule Crk
Uniprot
A0A2A4J7D4
A0A3S2LJZ6
A0A0N0PCV4
A0A194PVT4
A0A0L7LPM1
A0A182PA74
+ More
A0A087T469 A0A2M4ANU7 W5JCS8 A0A1Y1MAE5 A0A336KM49 A0A1Q3EZQ3 A0A2M4BW92 A0A182F565 A0A182KGM0 A0A1S4EKH5 Q7QG48 A0A1Q3EZ01 A0A182L5K4 A0A182TLI5 A0A182V0U0 A0A182X0E4 A0A182IE55 A0A1L8DLM6 A0A1B0CLT4 B0W5R9 A0A1S4J6E9 A0A182R117 A0A182M0X9 A0A182Y5V6 A0A182RGE0 K7IWQ3 A0A232EY17 A0A0P4VQ02 T1I0P2 A0A023F5B5 A0A0V0G7J0 A0A069DR22 A0A224XI25 A0A182IPR7 Q0IF64 A0A0A9WED1 A0A084VPI1 A0A182GSA0 A0A0P4ZQ04 A0A1B6E8K2 A0A1S3HHJ2 A0A1D1UTG6 A0A1E1X7T4 A0A1B6KTB1 T1JIX5 A0A1B6IWS6 A0A1B6G2X3 A0A3S4QUX1 E0VNA4 A0A0J7KK00 A0A1J1HTR5 A0A182NEG3 A0A226DGP1 A0A2P8Y829 A0A1B6D8E3 A0A3L8DAF9 E2AJ57 E2C5L7 A0A151WV37 E9J1P7 F4WKU4 A0A154PK49 A0A2A3EDF2 A0A2J7QYR6 A0A0Q9XF90 A0A2R5LMG8 A0A2H8TXT9 D6W7Z9 A0A131XES9 A0A224Z7A0 A0A131YRY2 A0A0M9ABN6 A0A0K8TKN9 A0A1I8P005 L7M1T1 A0A1W4WXR1 A0A2H8TUB2 A0A0K8UZH7 H9XVM7 T1PJ65 A0A0C9RDQ4 A0A0T6BG36 B4NHJ7 A0A1D2N376 A0A034WWY5 A0A2S2PN28 A0A0K8U2F3 A0A1A9WKY9 B3P9V1 A0A0J9S0Q9 B4IL88 Q9XYM0
A0A087T469 A0A2M4ANU7 W5JCS8 A0A1Y1MAE5 A0A336KM49 A0A1Q3EZQ3 A0A2M4BW92 A0A182F565 A0A182KGM0 A0A1S4EKH5 Q7QG48 A0A1Q3EZ01 A0A182L5K4 A0A182TLI5 A0A182V0U0 A0A182X0E4 A0A182IE55 A0A1L8DLM6 A0A1B0CLT4 B0W5R9 A0A1S4J6E9 A0A182R117 A0A182M0X9 A0A182Y5V6 A0A182RGE0 K7IWQ3 A0A232EY17 A0A0P4VQ02 T1I0P2 A0A023F5B5 A0A0V0G7J0 A0A069DR22 A0A224XI25 A0A182IPR7 Q0IF64 A0A0A9WED1 A0A084VPI1 A0A182GSA0 A0A0P4ZQ04 A0A1B6E8K2 A0A1S3HHJ2 A0A1D1UTG6 A0A1E1X7T4 A0A1B6KTB1 T1JIX5 A0A1B6IWS6 A0A1B6G2X3 A0A3S4QUX1 E0VNA4 A0A0J7KK00 A0A1J1HTR5 A0A182NEG3 A0A226DGP1 A0A2P8Y829 A0A1B6D8E3 A0A3L8DAF9 E2AJ57 E2C5L7 A0A151WV37 E9J1P7 F4WKU4 A0A154PK49 A0A2A3EDF2 A0A2J7QYR6 A0A0Q9XF90 A0A2R5LMG8 A0A2H8TXT9 D6W7Z9 A0A131XES9 A0A224Z7A0 A0A131YRY2 A0A0M9ABN6 A0A0K8TKN9 A0A1I8P005 L7M1T1 A0A1W4WXR1 A0A2H8TUB2 A0A0K8UZH7 H9XVM7 T1PJ65 A0A0C9RDQ4 A0A0T6BG36 B4NHJ7 A0A1D2N376 A0A034WWY5 A0A2S2PN28 A0A0K8U2F3 A0A1A9WKY9 B3P9V1 A0A0J9S0Q9 B4IL88 Q9XYM0
Pubmed
26354079
26227816
20920257
23761445
28004739
12364791
+ More
20966253 25244985 20075255 28648823 27129103 25474469 26334808 17510324 25401762 26823975 24438588 26483478 27649274 28503490 20566863 29403074 30249741 20798317 21282665 21719571 17994087 18362917 19820115 28049606 28797301 26830274 26369729 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 27289101 25348373 22936249 10072777
20966253 25244985 20075255 28648823 27129103 25474469 26334808 17510324 25401762 26823975 24438588 26483478 27649274 28503490 20566863 29403074 30249741 20798317 21282665 21719571 17994087 18362917 19820115 28049606 28797301 26830274 26369729 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 27289101 25348373 22936249 10072777
EMBL
NWSH01002557
PCG67995.1
RSAL01000085
RVE48335.1
KQ460466
KPJ14615.1
+ More
KQ459597 KPI95240.1 JTDY01000379 KOB77468.1 KK113338 KFM59908.1 GGFK01009145 MBW42466.1 ADMH02001881 ETN60685.1 GEZM01037757 JAV81958.1 UFQS01000668 UFQT01000668 SSX05974.1 SSX26331.1 GFDL01014271 JAV20774.1 GGFJ01008162 MBW57303.1 AAAB01008839 EAA05910.3 GFDL01014577 JAV20468.1 APCN01004569 GFDF01006715 JAV07369.1 AJWK01017609 DS231844 EDS35722.1 AXCN02000858 AXCM01000440 AAZX01007094 NNAY01001690 OXU23215.1 GDKW01002713 JAI53882.1 ACPB03016929 GBBI01002414 JAC16298.1 GECL01002049 JAP04075.1 GBGD01002564 JAC86325.1 GFTR01004291 JAW12135.1 CH477394 EAT41873.1 GBHO01040387 GBHO01040385 GBHO01040384 GBHO01040383 GBHO01040382 GBHO01014703 GBRD01003779 GDHC01010176 JAG03217.1 JAG03219.1 JAG03220.1 JAG03221.1 JAG03222.1 JAG28901.1 JAG62042.1 JAQ08453.1 ATLV01015007 KE524999 KFB39875.1 JXUM01005913 JXUM01084232 JXUM01084233 KQ560204 KXJ83839.1 GDIP01214002 GDIP01152781 GDIP01115860 JAJ09400.1 GEDC01003037 JAS34261.1 BDGG01000002 GAU92974.1 GFAC01003904 JAT95284.1 GEBQ01025302 GEBQ01011037 JAT14675.1 JAT28940.1 AFFK01016682 GECU01031934 GECU01016312 GECU01014205 JAS75772.1 JAS91394.1 JAS93501.1 GECZ01012997 JAS56772.1 NCKU01003150 RWS08075.1 DS235335 EEB14860.1 LBMM01006501 KMQ90561.1 CVRI01000021 CRK91464.1 LNIX01000020 OXA44014.1 PYGN01000816 PSN40416.1 GEDC01015366 JAS21932.1 QOIP01000011 RLU17132.1 GL439967 EFN66497.1 GL452770 EFN76830.1 KQ982708 KYQ51792.1 GL767674 EFZ13209.1 GL888206 EGI65186.1 KQ434931 KZC11834.1 KZ288288 PBC29322.1 NEVH01009083 PNF33725.1 CH933813 KRG07215.1 GGLE01006575 MBY10701.1 GFXV01007252 MBW19057.1 KQ971307 EFA11162.1 GEFH01004420 JAP64161.1 GFPF01011106 MAA22252.1 GEDV01007327 JAP81230.1 KQ435689 KOX81245.1 GDAI01002912 JAI14691.1 GACK01007966 JAA57068.1 GFXV01006058 MBW17863.1 GDHF01020245 JAI32069.1 AE014135 AFH06772.1 KA648160 AFP62789.1 GBYB01006455 JAG76222.1 LJIG01000734 KRT86206.1 CH964272 EDW83566.2 LJIJ01000258 ODM99737.1 GAKP01000258 GAKP01000257 JAC58695.1 GGMR01018165 MBY30784.1 GDHF01033081 GDHF01031633 GDHF01030801 GDHF01008551 GDHF01007068 JAI19233.1 JAI20681.1 JAI21513.1 JAI43763.1 JAI45246.1 CH954184 EDV45264.1 CM002916 KMZ07168.1 CH480867 EDW53719.1 AF112976 BT012456
KQ459597 KPI95240.1 JTDY01000379 KOB77468.1 KK113338 KFM59908.1 GGFK01009145 MBW42466.1 ADMH02001881 ETN60685.1 GEZM01037757 JAV81958.1 UFQS01000668 UFQT01000668 SSX05974.1 SSX26331.1 GFDL01014271 JAV20774.1 GGFJ01008162 MBW57303.1 AAAB01008839 EAA05910.3 GFDL01014577 JAV20468.1 APCN01004569 GFDF01006715 JAV07369.1 AJWK01017609 DS231844 EDS35722.1 AXCN02000858 AXCM01000440 AAZX01007094 NNAY01001690 OXU23215.1 GDKW01002713 JAI53882.1 ACPB03016929 GBBI01002414 JAC16298.1 GECL01002049 JAP04075.1 GBGD01002564 JAC86325.1 GFTR01004291 JAW12135.1 CH477394 EAT41873.1 GBHO01040387 GBHO01040385 GBHO01040384 GBHO01040383 GBHO01040382 GBHO01014703 GBRD01003779 GDHC01010176 JAG03217.1 JAG03219.1 JAG03220.1 JAG03221.1 JAG03222.1 JAG28901.1 JAG62042.1 JAQ08453.1 ATLV01015007 KE524999 KFB39875.1 JXUM01005913 JXUM01084232 JXUM01084233 KQ560204 KXJ83839.1 GDIP01214002 GDIP01152781 GDIP01115860 JAJ09400.1 GEDC01003037 JAS34261.1 BDGG01000002 GAU92974.1 GFAC01003904 JAT95284.1 GEBQ01025302 GEBQ01011037 JAT14675.1 JAT28940.1 AFFK01016682 GECU01031934 GECU01016312 GECU01014205 JAS75772.1 JAS91394.1 JAS93501.1 GECZ01012997 JAS56772.1 NCKU01003150 RWS08075.1 DS235335 EEB14860.1 LBMM01006501 KMQ90561.1 CVRI01000021 CRK91464.1 LNIX01000020 OXA44014.1 PYGN01000816 PSN40416.1 GEDC01015366 JAS21932.1 QOIP01000011 RLU17132.1 GL439967 EFN66497.1 GL452770 EFN76830.1 KQ982708 KYQ51792.1 GL767674 EFZ13209.1 GL888206 EGI65186.1 KQ434931 KZC11834.1 KZ288288 PBC29322.1 NEVH01009083 PNF33725.1 CH933813 KRG07215.1 GGLE01006575 MBY10701.1 GFXV01007252 MBW19057.1 KQ971307 EFA11162.1 GEFH01004420 JAP64161.1 GFPF01011106 MAA22252.1 GEDV01007327 JAP81230.1 KQ435689 KOX81245.1 GDAI01002912 JAI14691.1 GACK01007966 JAA57068.1 GFXV01006058 MBW17863.1 GDHF01020245 JAI32069.1 AE014135 AFH06772.1 KA648160 AFP62789.1 GBYB01006455 JAG76222.1 LJIG01000734 KRT86206.1 CH964272 EDW83566.2 LJIJ01000258 ODM99737.1 GAKP01000258 GAKP01000257 JAC58695.1 GGMR01018165 MBY30784.1 GDHF01033081 GDHF01031633 GDHF01030801 GDHF01008551 GDHF01007068 JAI19233.1 JAI20681.1 JAI21513.1 JAI43763.1 JAI45246.1 CH954184 EDV45264.1 CM002916 KMZ07168.1 CH480867 EDW53719.1 AF112976 BT012456
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
UP000075885
+ More
UP000054359 UP000000673 UP000069272 UP000075881 UP000079169 UP000007062 UP000075882 UP000075902 UP000075903 UP000076407 UP000075840 UP000092461 UP000002320 UP000075886 UP000075883 UP000076408 UP000075900 UP000002358 UP000215335 UP000015103 UP000075880 UP000008820 UP000030765 UP000069940 UP000249989 UP000085678 UP000186922 UP000285301 UP000009046 UP000036403 UP000183832 UP000075884 UP000198287 UP000245037 UP000279307 UP000000311 UP000008237 UP000075809 UP000007755 UP000076502 UP000242457 UP000235965 UP000009192 UP000007266 UP000053105 UP000095300 UP000192223 UP000000803 UP000095301 UP000007798 UP000094527 UP000091820 UP000008711 UP000001292
UP000054359 UP000000673 UP000069272 UP000075881 UP000079169 UP000007062 UP000075882 UP000075902 UP000075903 UP000076407 UP000075840 UP000092461 UP000002320 UP000075886 UP000075883 UP000076408 UP000075900 UP000002358 UP000215335 UP000015103 UP000075880 UP000008820 UP000030765 UP000069940 UP000249989 UP000085678 UP000186922 UP000285301 UP000009046 UP000036403 UP000183832 UP000075884 UP000198287 UP000245037 UP000279307 UP000000311 UP000008237 UP000075809 UP000007755 UP000076502 UP000242457 UP000235965 UP000009192 UP000007266 UP000053105 UP000095300 UP000192223 UP000000803 UP000095301 UP000007798 UP000094527 UP000091820 UP000008711 UP000001292
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J7D4
A0A3S2LJZ6
A0A0N0PCV4
A0A194PVT4
A0A0L7LPM1
A0A182PA74
+ More
A0A087T469 A0A2M4ANU7 W5JCS8 A0A1Y1MAE5 A0A336KM49 A0A1Q3EZQ3 A0A2M4BW92 A0A182F565 A0A182KGM0 A0A1S4EKH5 Q7QG48 A0A1Q3EZ01 A0A182L5K4 A0A182TLI5 A0A182V0U0 A0A182X0E4 A0A182IE55 A0A1L8DLM6 A0A1B0CLT4 B0W5R9 A0A1S4J6E9 A0A182R117 A0A182M0X9 A0A182Y5V6 A0A182RGE0 K7IWQ3 A0A232EY17 A0A0P4VQ02 T1I0P2 A0A023F5B5 A0A0V0G7J0 A0A069DR22 A0A224XI25 A0A182IPR7 Q0IF64 A0A0A9WED1 A0A084VPI1 A0A182GSA0 A0A0P4ZQ04 A0A1B6E8K2 A0A1S3HHJ2 A0A1D1UTG6 A0A1E1X7T4 A0A1B6KTB1 T1JIX5 A0A1B6IWS6 A0A1B6G2X3 A0A3S4QUX1 E0VNA4 A0A0J7KK00 A0A1J1HTR5 A0A182NEG3 A0A226DGP1 A0A2P8Y829 A0A1B6D8E3 A0A3L8DAF9 E2AJ57 E2C5L7 A0A151WV37 E9J1P7 F4WKU4 A0A154PK49 A0A2A3EDF2 A0A2J7QYR6 A0A0Q9XF90 A0A2R5LMG8 A0A2H8TXT9 D6W7Z9 A0A131XES9 A0A224Z7A0 A0A131YRY2 A0A0M9ABN6 A0A0K8TKN9 A0A1I8P005 L7M1T1 A0A1W4WXR1 A0A2H8TUB2 A0A0K8UZH7 H9XVM7 T1PJ65 A0A0C9RDQ4 A0A0T6BG36 B4NHJ7 A0A1D2N376 A0A034WWY5 A0A2S2PN28 A0A0K8U2F3 A0A1A9WKY9 B3P9V1 A0A0J9S0Q9 B4IL88 Q9XYM0
A0A087T469 A0A2M4ANU7 W5JCS8 A0A1Y1MAE5 A0A336KM49 A0A1Q3EZQ3 A0A2M4BW92 A0A182F565 A0A182KGM0 A0A1S4EKH5 Q7QG48 A0A1Q3EZ01 A0A182L5K4 A0A182TLI5 A0A182V0U0 A0A182X0E4 A0A182IE55 A0A1L8DLM6 A0A1B0CLT4 B0W5R9 A0A1S4J6E9 A0A182R117 A0A182M0X9 A0A182Y5V6 A0A182RGE0 K7IWQ3 A0A232EY17 A0A0P4VQ02 T1I0P2 A0A023F5B5 A0A0V0G7J0 A0A069DR22 A0A224XI25 A0A182IPR7 Q0IF64 A0A0A9WED1 A0A084VPI1 A0A182GSA0 A0A0P4ZQ04 A0A1B6E8K2 A0A1S3HHJ2 A0A1D1UTG6 A0A1E1X7T4 A0A1B6KTB1 T1JIX5 A0A1B6IWS6 A0A1B6G2X3 A0A3S4QUX1 E0VNA4 A0A0J7KK00 A0A1J1HTR5 A0A182NEG3 A0A226DGP1 A0A2P8Y829 A0A1B6D8E3 A0A3L8DAF9 E2AJ57 E2C5L7 A0A151WV37 E9J1P7 F4WKU4 A0A154PK49 A0A2A3EDF2 A0A2J7QYR6 A0A0Q9XF90 A0A2R5LMG8 A0A2H8TXT9 D6W7Z9 A0A131XES9 A0A224Z7A0 A0A131YRY2 A0A0M9ABN6 A0A0K8TKN9 A0A1I8P005 L7M1T1 A0A1W4WXR1 A0A2H8TUB2 A0A0K8UZH7 H9XVM7 T1PJ65 A0A0C9RDQ4 A0A0T6BG36 B4NHJ7 A0A1D2N376 A0A034WWY5 A0A2S2PN28 A0A0K8U2F3 A0A1A9WKY9 B3P9V1 A0A0J9S0Q9 B4IL88 Q9XYM0
PDB
2EYZ
E-value=1.0836e-20,
Score=240
Ontologies
GO
Topology
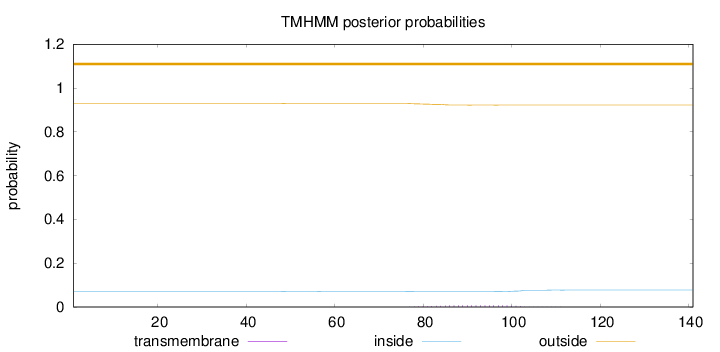
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16389
Exp number, first 60 AAs:
0.00088
Total prob of N-in:
0.07022
outside
1 - 141
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
