Gene
KWMTBOMO16657
Annotation
PREDICTED:_transcription_factor_EB-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.612
Sequence
CDS
ATGATTGCTTTCTATATTTTGTGGTCTTCTGTCCCCGGGGCACTGTTCTGGATGCTGGAGTTTTTAATACACGGACACACGCACAAGTTGAATCGCAAAGCACCTGGAGAAACGCTTTCCGCAGTTGAATATAAAGTTGTTGTTTTCGTTTGTTCCAGCCCGCCTACGTTTAAGACCCTGACGCCCACATCTCGCACACAACTGAAGCAGCAGTTGATGCGAGAACATGCCCAGGAGCAGCTGCGAAGAGAATCACAAAACCAACAAGCTGGACAATCCAAAGACAATGAAGAGCATAAAAAGAAGTCCTCGCCGACGGAGGTGCCTAGGATTACGCCACACGTCGAATTACCGCCACAAGTCCTACAGGTCCGAACAGTTCTGGAGAACCCCACCAGGTATCACGTGATACAGAAGCAGAAGAGTCAGGTACGACAGTACCTCAGCGAGTCCTTCCAACCACACACACAGGTCAGTGAAGTTTGGTCGTTTTTTTAA
Protein
MIAFYILWSSVPGALFWMLEFLIHGHTHKLNRKAPGETLSAVEYKVVVFVCSSPPTFKTLTPTSRTQLKQQLMREHAQEQLRRESQNQQAGQSKDNEEHKKKSSPTEVPRITPHVELPPQVLQVRTVLENPTRYHVIQKQKSQVRQYLSESFQPHTQVSEVWSFF
Summary
Uniprot
A0A3S2TFY2
A0A2A4KA52
A0A2W1BFJ0
A0A194QJM4
A0A151IG74
A0A1B6D8D0
+ More
A0A1B6CP76 A0A1B6CBE9 A0A1B6EA89 A0A1J1J5N2 A0A1J1J2K5 A0A1B6EHJ6 A0A0P8XYS8 A0A1B6IVF4 A0A1B6I793 A0A0P8XIE8 B3MZ93 A0A1B6KZF9 A0A1B6MU07 A0A1B6LBJ7 A0A0R1E7L8 C3KGP2 A0A026W1Q3 A0A0J9UT00 A0A0J9S308 B4IKS3 H9XVQ2 A0A2C9JTZ1 A0A1I8PF41
A0A1B6CP76 A0A1B6CBE9 A0A1B6EA89 A0A1J1J5N2 A0A1J1J2K5 A0A1B6EHJ6 A0A0P8XYS8 A0A1B6IVF4 A0A1B6I793 A0A0P8XIE8 B3MZ93 A0A1B6KZF9 A0A1B6MU07 A0A1B6LBJ7 A0A0R1E7L8 C3KGP2 A0A026W1Q3 A0A0J9UT00 A0A0J9S308 B4IKS3 H9XVQ2 A0A2C9JTZ1 A0A1I8PF41
Pubmed
EMBL
RSAL01000181
RVE44921.1
NWSH01000008
PCG80959.1
KZ150133
PZC73031.1
+ More
KQ458671 KPJ05584.1 KQ977726 KYN00269.1 GEDC01015350 JAS21948.1 GEDC01022107 JAS15191.1 GEDC01026501 JAS10797.1 GEDC01002450 JAS34848.1 CVRI01000066 CRL06193.1 CRL06194.1 GECZ01032398 JAS37371.1 CH902633 KPU74661.1 GECU01016787 JAS90919.1 GECU01024922 JAS82784.1 KPU74662.1 EDV33698.1 GEBQ01023134 JAT16843.1 GEBQ01000592 JAT39385.1 GEBQ01019073 JAT20904.1 CM000161 KRK05013.1 BT082107 AE014135 ACQ45345.1 AFH06796.1 KK107488 EZA49958.1 CM002916 KMZ07416.1 KMZ07415.1 CH480855 EDW52663.1 AFH06797.1
KQ458671 KPJ05584.1 KQ977726 KYN00269.1 GEDC01015350 JAS21948.1 GEDC01022107 JAS15191.1 GEDC01026501 JAS10797.1 GEDC01002450 JAS34848.1 CVRI01000066 CRL06193.1 CRL06194.1 GECZ01032398 JAS37371.1 CH902633 KPU74661.1 GECU01016787 JAS90919.1 GECU01024922 JAS82784.1 KPU74662.1 EDV33698.1 GEBQ01023134 JAT16843.1 GEBQ01000592 JAT39385.1 GEBQ01019073 JAT20904.1 CM000161 KRK05013.1 BT082107 AE014135 ACQ45345.1 AFH06796.1 KK107488 EZA49958.1 CM002916 KMZ07416.1 KMZ07415.1 CH480855 EDW52663.1 AFH06797.1
Proteomes
SUPFAM
SSF47459
SSF47459
Gene 3D
CDD
ProteinModelPortal
A0A3S2TFY2
A0A2A4KA52
A0A2W1BFJ0
A0A194QJM4
A0A151IG74
A0A1B6D8D0
+ More
A0A1B6CP76 A0A1B6CBE9 A0A1B6EA89 A0A1J1J5N2 A0A1J1J2K5 A0A1B6EHJ6 A0A0P8XYS8 A0A1B6IVF4 A0A1B6I793 A0A0P8XIE8 B3MZ93 A0A1B6KZF9 A0A1B6MU07 A0A1B6LBJ7 A0A0R1E7L8 C3KGP2 A0A026W1Q3 A0A0J9UT00 A0A0J9S308 B4IKS3 H9XVQ2 A0A2C9JTZ1 A0A1I8PF41
A0A1B6CP76 A0A1B6CBE9 A0A1B6EA89 A0A1J1J5N2 A0A1J1J2K5 A0A1B6EHJ6 A0A0P8XYS8 A0A1B6IVF4 A0A1B6I793 A0A0P8XIE8 B3MZ93 A0A1B6KZF9 A0A1B6MU07 A0A1B6LBJ7 A0A0R1E7L8 C3KGP2 A0A026W1Q3 A0A0J9UT00 A0A0J9S308 B4IKS3 H9XVQ2 A0A2C9JTZ1 A0A1I8PF41
Ontologies
GO
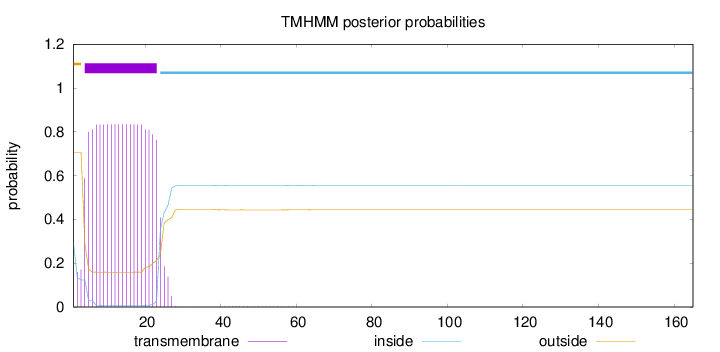
Topology
Length:
165
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.38139
Exp number, first 60 AAs:
17.37706
Total prob of N-in:
0.29400
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 23
inside
24 - 165
Population Genetic Test Statistics
Pi
17.799747
Theta
19.341149
Tajima's D
-0.477632
CLR
0.009072
CSRT
0.252337383130843
Interpretation
Uncertain
