Gene
KWMTBOMO16655
Annotation
PREDICTED:_uncharacterized_protein_LOC101743572_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.194 Extracellular Reliability : 1.466 Nuclear Reliability : 1.077
Sequence
CDS
ATGATCTCCGACGGAGATGAAGCCCAGGATACGCTTGTCACTAAACAGGGAATTCAAACGGATTTGCTGGACGGAGACGGAACGGTGGACCCTCGGTCCGTGGACCTCTCGCAGCTGCACCAGTCGCGCGTGGGCCTGATCAGCCTCGCGCTGATGCTCATACTGATCGTCTGCTTCCTGGCTTCTGAAACCCTAAGCGAATTCTGGAAGACTAAAAAAAATCCAACTCAAAAATAG
Protein
MISDGDEAQDTLVTKQGIQTDLLDGDGTVDPRSVDLSQLHQSRVGLISLALMLILIVCFLASETLSEFWKTKKNPTQK
Summary
Uniprot
EMBL
KQ459053
KPJ04206.1
GDQN01011454
JAT79600.1
KQ461181
KPJ07749.1
+ More
NWSH01002568 PCG67959.1 RSAL01000042 RVE50782.1 ODYU01006458 SOQ48315.1 JTDY01001123 KOB74807.1 GDIQ01014549 JAN80188.1 GDIQ01226398 JAK25327.1 GDIP01081049 JAM22666.1 GDIQ01227899 JAK23826.1 GDIQ01227898 JAK23827.1 GDIP01144877 JAJ78525.1 GDIP01116333 JAL87381.1 GDIP01012624 JAM91091.1 GDIQ01245850 GDIQ01209105 GDIQ01209104 GDIQ01191544 GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GL435242 EFN73588.1 GL443736 EFN61716.1 NEVH01012083 PNF30927.1 PNF30922.1
NWSH01002568 PCG67959.1 RSAL01000042 RVE50782.1 ODYU01006458 SOQ48315.1 JTDY01001123 KOB74807.1 GDIQ01014549 JAN80188.1 GDIQ01226398 JAK25327.1 GDIP01081049 JAM22666.1 GDIQ01227899 JAK23826.1 GDIQ01227898 JAK23827.1 GDIP01144877 JAJ78525.1 GDIP01116333 JAL87381.1 GDIP01012624 JAM91091.1 GDIQ01245850 GDIQ01209105 GDIQ01209104 GDIQ01191544 GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GL435242 EFN73588.1 GL443736 EFN61716.1 NEVH01012083 PNF30927.1 PNF30922.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Ontologies
Topology
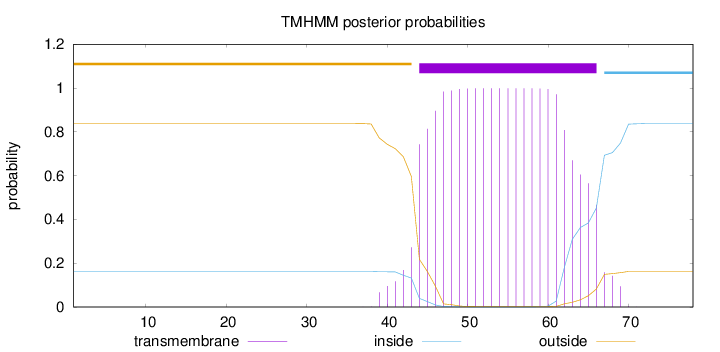
Length:
78
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.60174
Exp number, first 60 AAs:
17.1199
Total prob of N-in:
0.16205
POSSIBLE N-term signal
sequence
outside
1 - 43
TMhelix
44 - 66
inside
67 - 78
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
