Pre Gene Modal
BGIBMGA004595
Annotation
PREDICTED:_hexokinase-2-like_[Amyelois_transitella]
Full name
Phosphotransferase
Location in the cell
Cytoplasmic Reliability : 1.959
Sequence
CDS
ATGTCTGCTGCGGTAGAAAATCAGCGTACCGTCGAAATTGAGCTGGTGAATGGTCATGGCGGTGCGGACCCGCGAAAATGGGAGGACCTGTTTTTGCCCACCCCGCTCAAGCTGGAGAACAGGCTCAGAGCCCAGAAGATTGAAGCCCGTCTCAGCAAGTTAATGCTATGCGGCTCGACACTACGTAGAGTTGGAGAGGTGTTCGAGAAAGAAATCGAGGCGGGACTCCGACAACAGCCTTCGAGTTTGCAAATGGAAAATACTTATGTCCCAGAATTGCCTGACGGAACAGAGGAAGGCTCGTTCCTAGCGTTGGATTTGGGTGGTACAAATTTTCGGGTCCTGTTTTTGGAGTTGAAAGCCGGTCAATTGCTCCGTGAGAAAGTGAAACACTATCACATAAATGATGCCCTGCGACTGGGTCCCGGTGAAGATCTGTTTAACTTCTTAGCAGACTGCGTGCAGGATTTTTTGGTTTCACAATCTATTGAAAAAGAAACCTTCTCTTTGGGTTTCACATTCTCGTTTCCTATGAGGCAGCATTCGATATCCTCTGGAGAACTAATCACTTGGACGAAGAGCTTCAACTGCGGCGGAATGGACGGGGTTGATGTGGCTGCCCTGCTGCAGAGGTGCCTTAGCGCCAGGAGTCTGGATGTCACCGTGCGGGTGCTCCTCAATGATACAACTGGCACTCTGGTCGCTGGAGCTCATATCGATCGTAACGTTGGAATCGGCGTGATTCTGGGTACCGGCTCAAACGGGTGTTATATGGAGCGCGCGTCACGGGTCCTCCATTGGGAGCGTCCTCACTCCGCAGTGCACGATGTGTGCATCGACATCGAGTGGGGTGCCTTCGGCGACAACGGCTGCCTCGATTTCCTTCGCACAGAGTTCGACGACGAAGTGGACGCTAAGAGTCTGCTTGCCGCTTCTTTTACTTTCGAAAAGTACATAGGAGGCAAATATCTCGGGGACCTATTGACTTTTACTTTGAGTGCGTTGGCGCGAGATGGTCTGTTTCCAGCTGATCCTAATTTCCGTAAACTGCAGACATCGCATGTCAGTTTGTTCGAAGAAGAGAGCTGCGCCGGCGAAGTGTCTCGCACATTGTCAGCGCTCCGCGAAGCTTGCAATTCAGATTTGTTGAGCACAGCAGACGCCCTTGTAGCGCAGTATGTGGCGCACGTTATATCTAACCGAGCTGCCCAACTTGTGTCTGTTTGTATCTCGACATTACTCAAACGTATGGACAGGCCGTACGTGGCGGTGGCAGTTGACGGGTCCGTGTTCAAACATCACCCTCGTATCGCAAAACTCATGAATAAATATATAGCATTGCTAGCCCCAGCTCATCAATTTTCACTGATGATTGCTGAGGACGGTAGCGGCAAGGGCTCGGCCTTGACAGCGGCTATCGCAGCTCGTATGGCTACAAGAGGCGTGGCGAGGTTATAA
Protein
MSAAVENQRTVEIELVNGHGGADPRKWEDLFLPTPLKLENRLRAQKIEARLSKLMLCGSTLRRVGEVFEKEIEAGLRQQPSSLQMENTYVPELPDGTEEGSFLALDLGGTNFRVLFLELKAGQLLREKVKHYHINDALRLGPGEDLFNFLADCVQDFLVSQSIEKETFSLGFTFSFPMRQHSISSGELITWTKSFNCGGMDGVDVAALLQRCLSARSLDVTVRVLLNDTTGTLVAGAHIDRNVGIGVILGTGSNGCYMERASRVLHWERPHSAVHDVCIDIEWGAFGDNGCLDFLRTEFDDEVDAKSLLAASFTFEKYIGGKYLGDLLTFTLSALARDGLFPADPNFRKLQTSHVSLFEEESCAGEVSRTLSALREACNSDLLSTADALVAQYVAHVISNRAAQLVSVCISTLLKRMDRPYVAVAVDGSVFKHHPRIAKLMNKYIALLAPAHQFSLMIAEDGSGKGSALTAAIAARMATRGVARL
Summary
Similarity
Belongs to the hexokinase family.
Feature
chain Phosphotransferase
Uniprot
H9J505
A0A289ZNI8
A0A2W1B793
A0A2H1WNY8
A0A2A4JM95
A0A0N0P9H4
+ More
A0A0N1IBF8 A0A1S6KW35 A0A2J7QW31 A0A2J7QW41 A0A0C9PPV4 A0A088ADN5 V9IAF2 A0A2Z5RE13 A0A310SWU2 A0A1B6I221 A0A1B6HI11 A0A1B6MP98 A0A0M8ZVN1 E9H4L3 A0A2A3EMX7 A0A0P5FGT0 A0A3L8DVU6 A0A026WTR8 A0A0P5H201 A0A2P8Y546 A0A154PQC4 A0A195CJD9 A0A1B6CBM9 A0A151WTN1 A0A195DFC8 A0A195FTK8 A0A195B1Y3 A0A158NYM9 E2C3M9 E2ANK9 A0A067RFI2 A0A0P6BXT6 A0A1J1IDC2 A0A2R7WQI2 E0VSX4 A0A0J7L1N2 A0A0K8RD96 A0A1I8M2H4 A0A336LKM4 A0A1L8E278 A0A226D844 X2IYV8 A0A1E1XDM0 A0A1I8QEZ0 A0A0L7QY02 A0A0P4VIK7 A0A131XNR2 A0A1Z5KXD7 A0A023EZW9 A0A131Z378 A0A1D2M806 F4WXX5 A0A2R5L9E4 T1HIT8 A0A293MXA8 A0A224YYU3 A0A336LHR1 B1N691 A0A224XCI1 G3MQQ3 A0A0P6FY01 A0A0A9Z2D5 A0A0A9W080 A0A0A9Z2D9 A0A161N3T0 A0A2R5LNN3 A0A1B0CLD2 A0A293MVE0 A0A0A7DSW7 A0A023GJT0 E9J4T5 A0A087UWP9 A0A2P2I2H6 A0A0K2U132 A0A0P5YQR2 T1IWW6 A0A1S3JDN8 A0A336LEI4 A0A0A1WEY4 A0A238BUA6 A0A1S3I378 T1K546 A0A044TUF4 A0A182DZ92 A0A0N4T1I5 A0A0K0IMG9 T1J8Y0 A0A0R3S519 A0A2L2Y189 A0A131Y2F2 A0A0C9R2P6
A0A0N1IBF8 A0A1S6KW35 A0A2J7QW31 A0A2J7QW41 A0A0C9PPV4 A0A088ADN5 V9IAF2 A0A2Z5RE13 A0A310SWU2 A0A1B6I221 A0A1B6HI11 A0A1B6MP98 A0A0M8ZVN1 E9H4L3 A0A2A3EMX7 A0A0P5FGT0 A0A3L8DVU6 A0A026WTR8 A0A0P5H201 A0A2P8Y546 A0A154PQC4 A0A195CJD9 A0A1B6CBM9 A0A151WTN1 A0A195DFC8 A0A195FTK8 A0A195B1Y3 A0A158NYM9 E2C3M9 E2ANK9 A0A067RFI2 A0A0P6BXT6 A0A1J1IDC2 A0A2R7WQI2 E0VSX4 A0A0J7L1N2 A0A0K8RD96 A0A1I8M2H4 A0A336LKM4 A0A1L8E278 A0A226D844 X2IYV8 A0A1E1XDM0 A0A1I8QEZ0 A0A0L7QY02 A0A0P4VIK7 A0A131XNR2 A0A1Z5KXD7 A0A023EZW9 A0A131Z378 A0A1D2M806 F4WXX5 A0A2R5L9E4 T1HIT8 A0A293MXA8 A0A224YYU3 A0A336LHR1 B1N691 A0A224XCI1 G3MQQ3 A0A0P6FY01 A0A0A9Z2D5 A0A0A9W080 A0A0A9Z2D9 A0A161N3T0 A0A2R5LNN3 A0A1B0CLD2 A0A293MVE0 A0A0A7DSW7 A0A023GJT0 E9J4T5 A0A087UWP9 A0A2P2I2H6 A0A0K2U132 A0A0P5YQR2 T1IWW6 A0A1S3JDN8 A0A336LEI4 A0A0A1WEY4 A0A238BUA6 A0A1S3I378 T1K546 A0A044TUF4 A0A182DZ92 A0A0N4T1I5 A0A0K0IMG9 T1J8Y0 A0A0R3S519 A0A2L2Y189 A0A131Y2F2 A0A0C9R2P6
EC Number
2.7.1.-
Pubmed
EMBL
BABH01025763
KY624592
ATA67116.1
KZ150258
PZC71772.1
ODYU01009606
+ More
SOQ54154.1 NWSH01001113 PCG72553.1 KQ459444 KPJ00943.1 KQ460077 KPJ17860.1 KU556830 AQT25643.1 NEVH01009768 PNF32799.1 PNF32800.1 GBYB01003232 GBYB01003233 JAG72999.1 JAG73000.1 JR037958 JR037959 AEY58055.1 AEY58056.1 FX983725 BAX07224.1 KQ759820 OAD62753.1 GECU01026719 GECU01024766 JAS80987.1 JAS82940.1 GECU01033360 JAS74346.1 GEBQ01002200 JAT37777.1 KQ435824 KOX72065.1 GL732592 EFX73328.1 KZ288206 PBC33115.1 GDIQ01254748 GDIP01130446 JAJ96976.1 JAL73268.1 QOIP01000003 RLU24363.1 KK107107 EZA59407.1 GDIQ01255970 JAJ95754.1 PYGN01000927 PSN39304.1 KQ435037 KZC14116.1 KQ977696 KYN00552.1 GEDC01026598 JAS10700.1 KQ982748 KYQ51292.1 KQ980953 KYN11139.1 KQ981276 KYN43761.1 KQ976681 KYM78209.1 ADTU01004084 GL452328 EFN77452.1 GL441253 EFN64975.1 KK852667 KDR18923.1 GDIP01007882 JAM95833.1 CVRI01000047 CRK98269.1 KK855096 PTY20805.1 DS235758 EEB16480.1 LBMM01001396 KMQ96389.1 GADI01005339 JAA68469.1 UFQS01000005 UFQT01000005 SSW96904.1 SSX17291.1 GFDF01001402 JAV12682.1 LNIX01000028 OXA41712.1 KJ130470 AHN49716.1 GFAC01001899 JAT97289.1 KQ414697 KOC63493.1 GDKW01001937 JAI54658.1 GEFH01001370 JAP67211.1 GFJQ02007250 JAV99719.1 GBBI01003922 JAC14790.1 GEDV01002693 JAP85864.1 LJIJ01002930 ODM89085.1 GL888434 EGI60911.1 GGLE01001972 MBY06098.1 ACPB03009657 GFWV01019810 MAA44538.1 GFPF01008357 MAA19503.1 SSW96903.1 SSX17290.1 EF102106 ABO21409.1 GFTR01006290 JAW10136.1 JO844204 AEO35821.1 GDIQ01052520 JAN42217.1 GBHO01007624 GDHC01009975 JAG35980.1 JAQ08654.1 GBHO01045329 JAF98274.1 GBHO01007619 GBRD01016803 GBRD01016802 GDHC01013498 JAG35985.1 JAG49024.1 JAQ05131.1 GEMB01001743 JAS01422.1 GGLE01006939 MBY11065.1 AJWK01017091 GFWV01019748 MAA44476.1 KF951259 AIW65713.1 GBBM01001239 JAC34179.1 GL768119 EFZ12177.1 KK122037 KFM81788.1 IACF01002587 LAB68235.1 HACA01014582 CDW31943.1 GDIP01056117 JAM47598.1 JH431633 UFQS01001816 UFQT01001816 SSX12442.1 SSX31893.1 GBXI01016840 JAC97451.1 KZ269999 OZC08917.1 CAEY01001585 CMVM020000020 UYRW01000114 VDK63272.1 UZAD01000219 VDN83190.1 JH431968 IAAA01007901 IAAA01007902 IAAA01007903 IAAA01007904 LAA01933.1 GEFM01002739 JAP73057.1 GBZX01001450 JAG91290.1
SOQ54154.1 NWSH01001113 PCG72553.1 KQ459444 KPJ00943.1 KQ460077 KPJ17860.1 KU556830 AQT25643.1 NEVH01009768 PNF32799.1 PNF32800.1 GBYB01003232 GBYB01003233 JAG72999.1 JAG73000.1 JR037958 JR037959 AEY58055.1 AEY58056.1 FX983725 BAX07224.1 KQ759820 OAD62753.1 GECU01026719 GECU01024766 JAS80987.1 JAS82940.1 GECU01033360 JAS74346.1 GEBQ01002200 JAT37777.1 KQ435824 KOX72065.1 GL732592 EFX73328.1 KZ288206 PBC33115.1 GDIQ01254748 GDIP01130446 JAJ96976.1 JAL73268.1 QOIP01000003 RLU24363.1 KK107107 EZA59407.1 GDIQ01255970 JAJ95754.1 PYGN01000927 PSN39304.1 KQ435037 KZC14116.1 KQ977696 KYN00552.1 GEDC01026598 JAS10700.1 KQ982748 KYQ51292.1 KQ980953 KYN11139.1 KQ981276 KYN43761.1 KQ976681 KYM78209.1 ADTU01004084 GL452328 EFN77452.1 GL441253 EFN64975.1 KK852667 KDR18923.1 GDIP01007882 JAM95833.1 CVRI01000047 CRK98269.1 KK855096 PTY20805.1 DS235758 EEB16480.1 LBMM01001396 KMQ96389.1 GADI01005339 JAA68469.1 UFQS01000005 UFQT01000005 SSW96904.1 SSX17291.1 GFDF01001402 JAV12682.1 LNIX01000028 OXA41712.1 KJ130470 AHN49716.1 GFAC01001899 JAT97289.1 KQ414697 KOC63493.1 GDKW01001937 JAI54658.1 GEFH01001370 JAP67211.1 GFJQ02007250 JAV99719.1 GBBI01003922 JAC14790.1 GEDV01002693 JAP85864.1 LJIJ01002930 ODM89085.1 GL888434 EGI60911.1 GGLE01001972 MBY06098.1 ACPB03009657 GFWV01019810 MAA44538.1 GFPF01008357 MAA19503.1 SSW96903.1 SSX17290.1 EF102106 ABO21409.1 GFTR01006290 JAW10136.1 JO844204 AEO35821.1 GDIQ01052520 JAN42217.1 GBHO01007624 GDHC01009975 JAG35980.1 JAQ08654.1 GBHO01045329 JAF98274.1 GBHO01007619 GBRD01016803 GBRD01016802 GDHC01013498 JAG35985.1 JAG49024.1 JAQ05131.1 GEMB01001743 JAS01422.1 GGLE01006939 MBY11065.1 AJWK01017091 GFWV01019748 MAA44476.1 KF951259 AIW65713.1 GBBM01001239 JAC34179.1 GL768119 EFZ12177.1 KK122037 KFM81788.1 IACF01002587 LAB68235.1 HACA01014582 CDW31943.1 GDIP01056117 JAM47598.1 JH431633 UFQS01001816 UFQT01001816 SSX12442.1 SSX31893.1 GBXI01016840 JAC97451.1 KZ269999 OZC08917.1 CAEY01001585 CMVM020000020 UYRW01000114 VDK63272.1 UZAD01000219 VDN83190.1 JH431968 IAAA01007901 IAAA01007902 IAAA01007903 IAAA01007904 LAA01933.1 GEFM01002739 JAP73057.1 GBZX01001450 JAG91290.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000235965
UP000005203
+ More
UP000053105 UP000000305 UP000242457 UP000279307 UP000053097 UP000245037 UP000076502 UP000078542 UP000075809 UP000078492 UP000078541 UP000078540 UP000005205 UP000008237 UP000000311 UP000027135 UP000183832 UP000009046 UP000036403 UP000095301 UP000198287 UP000095300 UP000053825 UP000094527 UP000007755 UP000015103 UP000092461 UP000054359 UP000085678 UP000015104 UP000024404 UP000077448 UP000271087 UP000038020 UP000278627 UP000006672 UP000050640
UP000053105 UP000000305 UP000242457 UP000279307 UP000053097 UP000245037 UP000076502 UP000078542 UP000075809 UP000078492 UP000078541 UP000078540 UP000005205 UP000008237 UP000000311 UP000027135 UP000183832 UP000009046 UP000036403 UP000095301 UP000198287 UP000095300 UP000053825 UP000094527 UP000007755 UP000015103 UP000092461 UP000054359 UP000085678 UP000015104 UP000024404 UP000077448 UP000271087 UP000038020 UP000278627 UP000006672 UP000050640
PRIDE
Interpro
ProteinModelPortal
H9J505
A0A289ZNI8
A0A2W1B793
A0A2H1WNY8
A0A2A4JM95
A0A0N0P9H4
+ More
A0A0N1IBF8 A0A1S6KW35 A0A2J7QW31 A0A2J7QW41 A0A0C9PPV4 A0A088ADN5 V9IAF2 A0A2Z5RE13 A0A310SWU2 A0A1B6I221 A0A1B6HI11 A0A1B6MP98 A0A0M8ZVN1 E9H4L3 A0A2A3EMX7 A0A0P5FGT0 A0A3L8DVU6 A0A026WTR8 A0A0P5H201 A0A2P8Y546 A0A154PQC4 A0A195CJD9 A0A1B6CBM9 A0A151WTN1 A0A195DFC8 A0A195FTK8 A0A195B1Y3 A0A158NYM9 E2C3M9 E2ANK9 A0A067RFI2 A0A0P6BXT6 A0A1J1IDC2 A0A2R7WQI2 E0VSX4 A0A0J7L1N2 A0A0K8RD96 A0A1I8M2H4 A0A336LKM4 A0A1L8E278 A0A226D844 X2IYV8 A0A1E1XDM0 A0A1I8QEZ0 A0A0L7QY02 A0A0P4VIK7 A0A131XNR2 A0A1Z5KXD7 A0A023EZW9 A0A131Z378 A0A1D2M806 F4WXX5 A0A2R5L9E4 T1HIT8 A0A293MXA8 A0A224YYU3 A0A336LHR1 B1N691 A0A224XCI1 G3MQQ3 A0A0P6FY01 A0A0A9Z2D5 A0A0A9W080 A0A0A9Z2D9 A0A161N3T0 A0A2R5LNN3 A0A1B0CLD2 A0A293MVE0 A0A0A7DSW7 A0A023GJT0 E9J4T5 A0A087UWP9 A0A2P2I2H6 A0A0K2U132 A0A0P5YQR2 T1IWW6 A0A1S3JDN8 A0A336LEI4 A0A0A1WEY4 A0A238BUA6 A0A1S3I378 T1K546 A0A044TUF4 A0A182DZ92 A0A0N4T1I5 A0A0K0IMG9 T1J8Y0 A0A0R3S519 A0A2L2Y189 A0A131Y2F2 A0A0C9R2P6
A0A0N1IBF8 A0A1S6KW35 A0A2J7QW31 A0A2J7QW41 A0A0C9PPV4 A0A088ADN5 V9IAF2 A0A2Z5RE13 A0A310SWU2 A0A1B6I221 A0A1B6HI11 A0A1B6MP98 A0A0M8ZVN1 E9H4L3 A0A2A3EMX7 A0A0P5FGT0 A0A3L8DVU6 A0A026WTR8 A0A0P5H201 A0A2P8Y546 A0A154PQC4 A0A195CJD9 A0A1B6CBM9 A0A151WTN1 A0A195DFC8 A0A195FTK8 A0A195B1Y3 A0A158NYM9 E2C3M9 E2ANK9 A0A067RFI2 A0A0P6BXT6 A0A1J1IDC2 A0A2R7WQI2 E0VSX4 A0A0J7L1N2 A0A0K8RD96 A0A1I8M2H4 A0A336LKM4 A0A1L8E278 A0A226D844 X2IYV8 A0A1E1XDM0 A0A1I8QEZ0 A0A0L7QY02 A0A0P4VIK7 A0A131XNR2 A0A1Z5KXD7 A0A023EZW9 A0A131Z378 A0A1D2M806 F4WXX5 A0A2R5L9E4 T1HIT8 A0A293MXA8 A0A224YYU3 A0A336LHR1 B1N691 A0A224XCI1 G3MQQ3 A0A0P6FY01 A0A0A9Z2D5 A0A0A9W080 A0A0A9Z2D9 A0A161N3T0 A0A2R5LNN3 A0A1B0CLD2 A0A293MVE0 A0A0A7DSW7 A0A023GJT0 E9J4T5 A0A087UWP9 A0A2P2I2H6 A0A0K2U132 A0A0P5YQR2 T1IWW6 A0A1S3JDN8 A0A336LEI4 A0A0A1WEY4 A0A238BUA6 A0A1S3I378 T1K546 A0A044TUF4 A0A182DZ92 A0A0N4T1I5 A0A0K0IMG9 T1J8Y0 A0A0R3S519 A0A2L2Y189 A0A131Y2F2 A0A0C9R2P6
PDB
5HG1
E-value=6.60479e-72,
Score=689
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
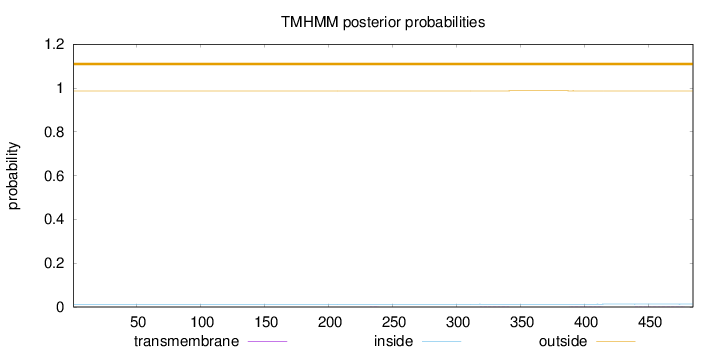
Topology
Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03945
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.01313
outside
1 - 485
Population Genetic Test Statistics
Pi
15.357117
Theta
19.911872
Tajima's D
-1.315722
CLR
0.071129
CSRT
0.0813959302034898
Interpretation
Uncertain
