Gene
KWMTBOMO16614
Annotation
PREDICTED:_TBC1_domain_family_member_20?_partial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.744
Sequence
CDS
ATGGAATCCAATAATAAAGACAGTGAAGTTATAGATAAAAGTGTTATGAATGGTGAATGTTCACCAGCAGAGAGTATTCATCCGACAAATTTGAATAAAACGAAAAATATTGATGTCATTTCGACACCCTCCGAATTAAAATTCGATAATGAGCCGGAGGAGGAAGACCCCGAGATTGTAGAAAAGAGGAAAGAGATTGAGGAATGTCTCTCTGATGTCGATTCTGTTGATGTGGAAAAATGGAAGAATCTAGCCAGAAGCAAAGGAGGACTTATCTGTGATGAATACAGAAGAAGAATTTGGCCTCTGCTTGTTGGTGTTACAAAAGAGGAAATGACTGAAGCTCCATCATTGGATGAGTTATCAACGCATCCAGAATATAACCAGGTGGTGCTAGATGTGAACAGGTCTTTGAAAAGATTTCCTCCTGGCATTCCATACGAACAAAGAGTGGCCCTTCAGGACCAGCTGACAGTATTAATACTCAGGGTCATCATAAAATATCCACATTTAAAATATTACCAGTTTGAGAGAGCGCTTCGTGGTTTATGA
Protein
MESNNKDSEVIDKSVMNGECSPAESIHPTNLNKTKNIDVISTPSELKFDNEPEEEDPEIVEKRKEIEECLSDVDSVDVEKWKNLARSKGGLICDEYRRRIWPLLVGVTKEEMTEAPSLDELSTHPEYNQVVLDVNRSLKRFPPGIPYEQRVALQDQLTVLILRVIIKYPHLKYYQFERALRGL
Summary
Uniprot
Pubmed
EMBL
AGBW02013812
OWR42722.1
BABH01034447
ODYU01005372
SOQ46143.1
NWSH01000050
+ More
PCG80182.1 KQ461191 KPJ07097.1 GDQN01008320 JAT82734.1 KQ458860 KPJ04759.1 JTDY01000935 KOB75370.1 AXCM01002003 AXCN02000803 CH477219 EAT47398.1 KB631676 ERL85319.1 UFQT01000055 SSX19093.1 CVRI01000038 CRK94380.1 GEZM01009504 JAV94546.1 LJIG01022744 KRT78943.1 GBHO01044157 GBRD01000514 JAF99446.1 JAG65307.1 GDHC01014086 JAQ04543.1 KQ971343 EFA03356.2 GEDC01026199 GEDC01024939 GEDC01018723 JAS11099.1 JAS12359.1 JAS18575.1 GFTR01006042 JAW10384.1 GECL01000412 JAP05712.1 GBGD01001607 JAC87282.1
PCG80182.1 KQ461191 KPJ07097.1 GDQN01008320 JAT82734.1 KQ458860 KPJ04759.1 JTDY01000935 KOB75370.1 AXCM01002003 AXCN02000803 CH477219 EAT47398.1 KB631676 ERL85319.1 UFQT01000055 SSX19093.1 CVRI01000038 CRK94380.1 GEZM01009504 JAV94546.1 LJIG01022744 KRT78943.1 GBHO01044157 GBRD01000514 JAF99446.1 JAG65307.1 GDHC01014086 JAQ04543.1 KQ971343 EFA03356.2 GEDC01026199 GEDC01024939 GEDC01018723 JAS11099.1 JAS12359.1 JAS18575.1 GFTR01006042 JAW10384.1 GECL01000412 JAP05712.1 GBGD01001607 JAC87282.1
Proteomes
PRIDE
Pfam
PF00566 RabGAP-TBC
SUPFAM
SSF47923
SSF47923
ProteinModelPortal
PDB
4HLQ
E-value=2.51398e-16,
Score=204
Ontologies
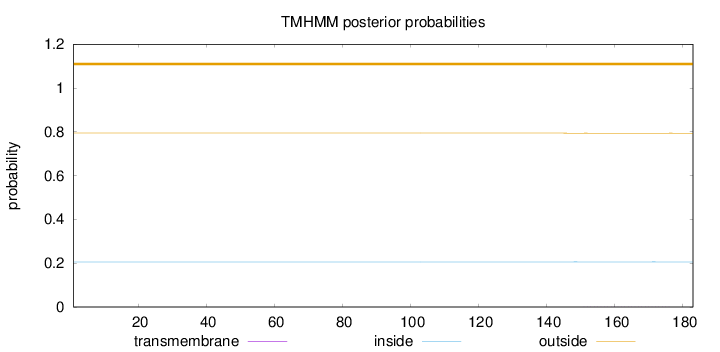
Topology
Length:
183
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01105
Exp number, first 60 AAs:
0
Total prob of N-in:
0.20588
outside
1 - 183
Population Genetic Test Statistics
Pi
12.657412
Theta
14.302821
Tajima's D
-0.344909
CLR
0
CSRT
0.285135743212839
Interpretation
Uncertain
