Gene
KWMTBOMO16582
Pre Gene Modal
BGIBMGA005583
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC106142549_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.323
Sequence
CDS
ATGACTACGAATGGGAATAGTGCCAACTACGCACCCCTCAAGCAAATACTATCGGAATCAGAGTCAGAAGATGACAACAAGTTGGAAAATGCAGTTCAAGCCGGCGACGGAAGAAATATGGATTTCAACACCGGTCTGTTGGTATCTAGGAATAGTATGAGTGACATGGACGATTTTAACAGCAACCTTAACACTATGGAGAGCACCATGGATAATGTAAGCTTCCTACAATCCGATATGTCCAGCAAATTATCAACACCACGAAGATTCGCGTTCATCACTTCAATTCTTCTGTGTATTATTGTCATAATCGTGTTCTTGTGGGGTATACCTTGTTCTGATTTCGGTAGCTGCCCAGGTGCTGAGTGGCAGGACAAAACAACTAGCTGGGAGTTTCCTTACGATGAAGTTGAATTGTCAGGAGCGGTGCAGGTCGTTGACGGTGCAATCCCGTTTACTAAAAACTTAATTTTTATTTATCGTGGGAACCACATGAGAAAGGATTCACCTAATGATGATAATGTGAATGGAGTATTGTTGATTGTTGGAAACACAGGTAAAGTGGGATGGTACACTCGTGAGAGTAGAATACCAACTGAAATCAACTGCCATTTGATTGATGTAAACAATGACAAGATTAAAGATTGTATTGTCTCTGGTTCAGAAGGTTTACTTGCTGCTATTAGCCCTCTTGCAGGTACTTACTATTGGTACATGCATAAACAAGGTAAAGTACTTAGCAATATAGCTGCAATTGATTTCCCGATTGTAATGAAAGATATGGATAAAGATAATGTGAGCGATCTGCTTACAGTTGCTACTGTTTACCCAAATACTAATCATAATTCACTACTAGTCATATCAGGTGCAACAGGTAACATCATTGGAGAACCTCTTGTAATGAATGATTGCTTGACAGTTAAATTGATTTCAGTCTCTGAAACAATCTCATATATTTGCAAGAATGGTGCTACAGAAGCTGTAAGGGACAAAGAATATTCAGTGTTGTATAAAAAACTGGTGACCGTTGAGCATGCAAATCATACTAAGCATGCGAAGCCAATTAATAAAAAAGTTAACATCAATTTTAAGAAAAGTATTGGAAATACCCGTGTTCTATATTCAAACGGTCCAGGCAAACTAATTGTTGAAAATTCTGGAGAATGCCCTGATTCATGTAAAGTGAATTTAACTTTATTGCTGGACAGAAACGGCAGCAGTACAGTCAGCTGGGAGTACACAGCAAATTATGTGTTTGCAATGAAACCAAGTACTTTTGCTTTCGCCAATTCTATACGAGGTTTTGTATTGAAATTATGA
Protein
MTTNGNSANYAPLKQILSESESEDDNKLENAVQAGDGRNMDFNTGLLVSRNSMSDMDDFNSNLNTMESTMDNVSFLQSDMSSKLSTPRRFAFITSILLCIIVIIVFLWGIPCSDFGSCPGAEWQDKTTSWEFPYDEVELSGAVQVVDGAIPFTKNLIFIYRGNHMRKDSPNDDNVNGVLLIVGNTGKVGWYTRESRIPTEINCHLIDVNNDKIKDCIVSGSEGLLAAISPLAGTYYWYMHKQGKVLSNIAAIDFPIVMKDMDKDNVSDLLTVATVYPNTNHNSLLVISGATGNIIGEPLVMNDCLTVKLISVSETISYICKNGATEAVRDKEYSVLYKKLVTVEHANHTKHAKPINKKVNINFKKSIGNTRVLYSNGPGKLIVENSGECPDSCKVNLTLLLDRNGSSTVSWEYTANYVFAMKPSTFAFANSIRGFVLKL
Summary
Uniprot
H9J7U1
A0A3S2NI04
A0A1E1W362
A0A2H1W6R5
A0A2A4K026
A0A0L7LJS8
+ More
A0A194QGU2 A0A212EPI3 A0A194QSH0 A0A182GVD9 A0A182G5Y0 Q16T59 Q17IS1 A0A0L0CK28 B0WKC8 B0X9J8 A0A182FH08 W5JBP9 T1PA89 A0A2M4CGD2 A0A1I8NCE8 A0A1I8P3V7 A0A1Y1KLW6 A0A182KBR3 A0A1B0GF58 A0A1B0BBN7 A0A3B0J4S7 A0A1A9UYH8 Q7Q6A8 A0A1A9YL06 A0A182X707 A0A182I1X7 Q2M148 A0A182SPF7 A0A182WKQ6 A0A182KPM9 A0A182LV19 A0A182IKM3 A0A182YD23 A0A1B0A0E1 A0A182V5C4 A0A182NIN5 A0A182QRA7 A0A182TKU6 A0A182PH61 A0A182RZQ1 B4GUN7 A0A0K8WEQ3 A0A034V1F9 A0A1B0CQA3 A0A1A9W2B7 W8B1Y7 B4N3X0 B3M6X0 A0A2J7RER1 B4L9G1 A0A1W4UQL7 B4IAG2 A0A067QXM0 B4QJ77 A0A0R1EF50 A0A0T6AZE6 Q9VPA9 B4LB62 B4PEP5 B3NIS3 B4J0Y9 A0A0M5IYH9 A0A1B6L6I5 A0A1B6CIX4 U4UTE3 A0A1B6E3T2 A0A1B6IWN8 A0A1B6DH06 A0A1B6FAQ1 A0A026WJZ1 T1GD13 A0A158NUE4 F4WNK1 T1I0F8 A0A069DUT3 A0A0V0G721 A0A0P4VMP6 K7IN25 A0A232F842 A0A151WGU0
A0A194QGU2 A0A212EPI3 A0A194QSH0 A0A182GVD9 A0A182G5Y0 Q16T59 Q17IS1 A0A0L0CK28 B0WKC8 B0X9J8 A0A182FH08 W5JBP9 T1PA89 A0A2M4CGD2 A0A1I8NCE8 A0A1I8P3V7 A0A1Y1KLW6 A0A182KBR3 A0A1B0GF58 A0A1B0BBN7 A0A3B0J4S7 A0A1A9UYH8 Q7Q6A8 A0A1A9YL06 A0A182X707 A0A182I1X7 Q2M148 A0A182SPF7 A0A182WKQ6 A0A182KPM9 A0A182LV19 A0A182IKM3 A0A182YD23 A0A1B0A0E1 A0A182V5C4 A0A182NIN5 A0A182QRA7 A0A182TKU6 A0A182PH61 A0A182RZQ1 B4GUN7 A0A0K8WEQ3 A0A034V1F9 A0A1B0CQA3 A0A1A9W2B7 W8B1Y7 B4N3X0 B3M6X0 A0A2J7RER1 B4L9G1 A0A1W4UQL7 B4IAG2 A0A067QXM0 B4QJ77 A0A0R1EF50 A0A0T6AZE6 Q9VPA9 B4LB62 B4PEP5 B3NIS3 B4J0Y9 A0A0M5IYH9 A0A1B6L6I5 A0A1B6CIX4 U4UTE3 A0A1B6E3T2 A0A1B6IWN8 A0A1B6DH06 A0A1B6FAQ1 A0A026WJZ1 T1GD13 A0A158NUE4 F4WNK1 T1I0F8 A0A069DUT3 A0A0V0G721 A0A0P4VMP6 K7IN25 A0A232F842 A0A151WGU0
Pubmed
19121390
26227816
26354079
22118469
26483478
17510324
+ More
26108605 20920257 23761445 25315136 28004739 12364791 14747013 17210077 15632085 17994087 20966253 25244985 25348373 24495485 24845553 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 24508170 30249741 21347285 21719571 26334808 27129103 20075255 28648823
26108605 20920257 23761445 25315136 28004739 12364791 14747013 17210077 15632085 17994087 20966253 25244985 25348373 24495485 24845553 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 24508170 30249741 21347285 21719571 26334808 27129103 20075255 28648823
EMBL
BABH01020142
RSAL01000090
RVE48067.1
GDQN01009644
JAT81410.1
ODYU01006570
+ More
SOQ48522.1 NWSH01000358 PCG77020.1 JTDY01000867 KOB75619.1 KQ459232 KPJ02666.1 AGBW02013457 OWR43389.1 KQ461155 KPJ08264.1 JXUM01090691 KQ563858 KXJ73158.1 JXUM01044144 KQ561389 KXJ78741.1 CH477659 EAT37648.1 CH477237 EAT46567.1 JRES01000378 KNC31829.1 DS231969 EDS29719.1 DS232539 EDS43167.1 ADMH02001858 ETN60833.1 KA644853 AFP59482.1 GGFL01000196 MBW64374.1 GEZM01086012 JAV59837.1 CCAG010012761 JXJN01011541 OUUW01000002 SPP76874.1 AAAB01008960 EAA11488.4 APCN01000100 CH379069 EAL30728.2 AXCM01009619 AXCN02000418 CH479191 EDW26320.1 GDHF01002673 JAI49641.1 GAKP01022613 JAC36339.1 AJWK01023331 GAMC01015512 JAB91043.1 CH964095 EDW79325.2 CH902618 EDV39806.1 NEVH01004957 PNF39307.1 CH933816 EDW17336.2 CH480826 EDW44275.1 KK853239 KDR09496.1 CM000363 CM002912 EDX11294.1 KMZ00871.1 CH891925 KRK05367.1 LJIG01022486 KRT80324.1 AE014296 AY058558 AAF51646.1 AAL13787.1 CH940647 EDW68626.1 CM000159 EDW95119.1 CH954178 EDV52569.1 CH916366 EDV95810.1 CP012525 ALC43548.1 GEBQ01025231 GEBQ01020645 JAT14746.1 JAT19332.1 GEDC01023946 JAS13352.1 KB632355 ERL93390.1 GEDC01004727 JAS32571.1 GECU01016361 JAS91345.1 GEDC01012334 JAS24964.1 GECZ01029403 GECZ01022472 JAS40366.1 JAS47297.1 KK107167 QOIP01000010 EZA56330.1 RLU17804.1 CAQQ02043020 ADTU01002848 GL888237 EGI64194.1 ACPB03006600 GBGD01001313 JAC87576.1 GECL01002278 JAP03846.1 GDKW01002993 JAI53602.1 NNAY01000681 OXU27004.1 KQ983150 KYQ47044.1
SOQ48522.1 NWSH01000358 PCG77020.1 JTDY01000867 KOB75619.1 KQ459232 KPJ02666.1 AGBW02013457 OWR43389.1 KQ461155 KPJ08264.1 JXUM01090691 KQ563858 KXJ73158.1 JXUM01044144 KQ561389 KXJ78741.1 CH477659 EAT37648.1 CH477237 EAT46567.1 JRES01000378 KNC31829.1 DS231969 EDS29719.1 DS232539 EDS43167.1 ADMH02001858 ETN60833.1 KA644853 AFP59482.1 GGFL01000196 MBW64374.1 GEZM01086012 JAV59837.1 CCAG010012761 JXJN01011541 OUUW01000002 SPP76874.1 AAAB01008960 EAA11488.4 APCN01000100 CH379069 EAL30728.2 AXCM01009619 AXCN02000418 CH479191 EDW26320.1 GDHF01002673 JAI49641.1 GAKP01022613 JAC36339.1 AJWK01023331 GAMC01015512 JAB91043.1 CH964095 EDW79325.2 CH902618 EDV39806.1 NEVH01004957 PNF39307.1 CH933816 EDW17336.2 CH480826 EDW44275.1 KK853239 KDR09496.1 CM000363 CM002912 EDX11294.1 KMZ00871.1 CH891925 KRK05367.1 LJIG01022486 KRT80324.1 AE014296 AY058558 AAF51646.1 AAL13787.1 CH940647 EDW68626.1 CM000159 EDW95119.1 CH954178 EDV52569.1 CH916366 EDV95810.1 CP012525 ALC43548.1 GEBQ01025231 GEBQ01020645 JAT14746.1 JAT19332.1 GEDC01023946 JAS13352.1 KB632355 ERL93390.1 GEDC01004727 JAS32571.1 GECU01016361 JAS91345.1 GEDC01012334 JAS24964.1 GECZ01029403 GECZ01022472 JAS40366.1 JAS47297.1 KK107167 QOIP01000010 EZA56330.1 RLU17804.1 CAQQ02043020 ADTU01002848 GL888237 EGI64194.1 ACPB03006600 GBGD01001313 JAC87576.1 GECL01002278 JAP03846.1 GDKW01002993 JAI53602.1 NNAY01000681 OXU27004.1 KQ983150 KYQ47044.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000053268
UP000007151
+ More
UP000053240 UP000069940 UP000249989 UP000008820 UP000037069 UP000002320 UP000069272 UP000000673 UP000095301 UP000095300 UP000075881 UP000092444 UP000092460 UP000268350 UP000078200 UP000007062 UP000092443 UP000076407 UP000075840 UP000001819 UP000075901 UP000075920 UP000075882 UP000075883 UP000075880 UP000076408 UP000092445 UP000075903 UP000075884 UP000075886 UP000075902 UP000075885 UP000075900 UP000008744 UP000092461 UP000091820 UP000007798 UP000007801 UP000235965 UP000009192 UP000192221 UP000001292 UP000027135 UP000000304 UP000002282 UP000000803 UP000008792 UP000008711 UP000001070 UP000092553 UP000030742 UP000053097 UP000279307 UP000015102 UP000005205 UP000007755 UP000015103 UP000002358 UP000215335 UP000075809
UP000053240 UP000069940 UP000249989 UP000008820 UP000037069 UP000002320 UP000069272 UP000000673 UP000095301 UP000095300 UP000075881 UP000092444 UP000092460 UP000268350 UP000078200 UP000007062 UP000092443 UP000076407 UP000075840 UP000001819 UP000075901 UP000075920 UP000075882 UP000075883 UP000075880 UP000076408 UP000092445 UP000075903 UP000075884 UP000075886 UP000075902 UP000075885 UP000075900 UP000008744 UP000092461 UP000091820 UP000007798 UP000007801 UP000235965 UP000009192 UP000192221 UP000001292 UP000027135 UP000000304 UP000002282 UP000000803 UP000008792 UP000008711 UP000001070 UP000092553 UP000030742 UP000053097 UP000279307 UP000015102 UP000005205 UP000007755 UP000015103 UP000002358 UP000215335 UP000075809
Pfam
PF03102 NeuB
Interpro
SUPFAM
SSF50998
SSF50998
Gene 3D
ProteinModelPortal
H9J7U1
A0A3S2NI04
A0A1E1W362
A0A2H1W6R5
A0A2A4K026
A0A0L7LJS8
+ More
A0A194QGU2 A0A212EPI3 A0A194QSH0 A0A182GVD9 A0A182G5Y0 Q16T59 Q17IS1 A0A0L0CK28 B0WKC8 B0X9J8 A0A182FH08 W5JBP9 T1PA89 A0A2M4CGD2 A0A1I8NCE8 A0A1I8P3V7 A0A1Y1KLW6 A0A182KBR3 A0A1B0GF58 A0A1B0BBN7 A0A3B0J4S7 A0A1A9UYH8 Q7Q6A8 A0A1A9YL06 A0A182X707 A0A182I1X7 Q2M148 A0A182SPF7 A0A182WKQ6 A0A182KPM9 A0A182LV19 A0A182IKM3 A0A182YD23 A0A1B0A0E1 A0A182V5C4 A0A182NIN5 A0A182QRA7 A0A182TKU6 A0A182PH61 A0A182RZQ1 B4GUN7 A0A0K8WEQ3 A0A034V1F9 A0A1B0CQA3 A0A1A9W2B7 W8B1Y7 B4N3X0 B3M6X0 A0A2J7RER1 B4L9G1 A0A1W4UQL7 B4IAG2 A0A067QXM0 B4QJ77 A0A0R1EF50 A0A0T6AZE6 Q9VPA9 B4LB62 B4PEP5 B3NIS3 B4J0Y9 A0A0M5IYH9 A0A1B6L6I5 A0A1B6CIX4 U4UTE3 A0A1B6E3T2 A0A1B6IWN8 A0A1B6DH06 A0A1B6FAQ1 A0A026WJZ1 T1GD13 A0A158NUE4 F4WNK1 T1I0F8 A0A069DUT3 A0A0V0G721 A0A0P4VMP6 K7IN25 A0A232F842 A0A151WGU0
A0A194QGU2 A0A212EPI3 A0A194QSH0 A0A182GVD9 A0A182G5Y0 Q16T59 Q17IS1 A0A0L0CK28 B0WKC8 B0X9J8 A0A182FH08 W5JBP9 T1PA89 A0A2M4CGD2 A0A1I8NCE8 A0A1I8P3V7 A0A1Y1KLW6 A0A182KBR3 A0A1B0GF58 A0A1B0BBN7 A0A3B0J4S7 A0A1A9UYH8 Q7Q6A8 A0A1A9YL06 A0A182X707 A0A182I1X7 Q2M148 A0A182SPF7 A0A182WKQ6 A0A182KPM9 A0A182LV19 A0A182IKM3 A0A182YD23 A0A1B0A0E1 A0A182V5C4 A0A182NIN5 A0A182QRA7 A0A182TKU6 A0A182PH61 A0A182RZQ1 B4GUN7 A0A0K8WEQ3 A0A034V1F9 A0A1B0CQA3 A0A1A9W2B7 W8B1Y7 B4N3X0 B3M6X0 A0A2J7RER1 B4L9G1 A0A1W4UQL7 B4IAG2 A0A067QXM0 B4QJ77 A0A0R1EF50 A0A0T6AZE6 Q9VPA9 B4LB62 B4PEP5 B3NIS3 B4J0Y9 A0A0M5IYH9 A0A1B6L6I5 A0A1B6CIX4 U4UTE3 A0A1B6E3T2 A0A1B6IWN8 A0A1B6DH06 A0A1B6FAQ1 A0A026WJZ1 T1GD13 A0A158NUE4 F4WNK1 T1I0F8 A0A069DUT3 A0A0V0G721 A0A0P4VMP6 K7IN25 A0A232F842 A0A151WGU0
Ontologies
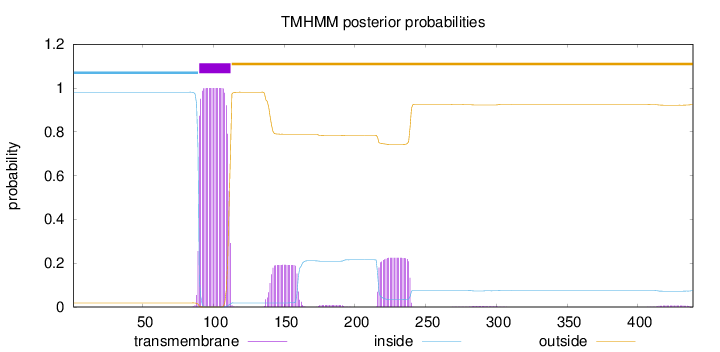
Topology
Length:
439
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
30.99822
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98118
inside
1 - 89
TMhelix
90 - 112
outside
113 - 439
Population Genetic Test Statistics
Pi
33.08801
Theta
41.318289
Tajima's D
-0.620219
CLR
0
CSRT
0.214839258037098
Interpretation
Uncertain
