Gene
KWMTBOMO16581
Pre Gene Modal
BGIBMGA005582
Annotation
SOCS2-12_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.114 Nuclear Reliability : 1.131
Sequence
CDS
ATGACTCTTGCAGCGCGAATTACCCCACAAAATGCGTGCTTGGAAGTAGGTGTGCCTCTGGGTGGCTGGTCGTCAGGTGTGGCATGCTGTCCGAATTGCAGGCACCAATTGCGAGTATCACTCGCGCCGGCGTCGGGCAAGTGTCACGCTTTGCCGTCGCCGCACACGCCGCCTTTCACCGTCCCCCCAACGTATTGGCCACATTCGCCTTTGTACGCGCCGCCATCACCGATCGTTCCGGCGCCTTACAATGATGAACTGAAGCGGCTTGCTGATACCCTCAGTGCGCTTAGGCTCTCTGGGTGGTACTACGATACTTTGAATTGGCAGGGAGCGAGAACATTGTTAAAAGACGCCAGTGTCGGAGCTTTTGTCGTGAGAGACTCTGGAGATAGAAACTTTATATTCTCACTATCGGTGCAAACGGAAAGGGGCCCTACATCCGTGAGGCTTCACTACGAACAAGGATTCTTCAGGTTAGATTGTGACAGACCGTTGGCTAGGTACATGCCCCGGTTCAGATGCGTGGTGGAAATGGTGCAGCAGTACAGACGGGCGGGACAACGCGGTGCGGCCGGTACGGTGTGGGTGGACCGCGAGGGGTGTCCGCATTCGCCGGTGTTACTCACCCATCCACTAAGGAAGAGCCCCCCATCTCTCATGCACGCGGCCCGTCTCGCTGTACACAAAGCCCTCGACTCCAACCCCCTCACTCCCAAGCTCTGGTGCGCTCCTAAACACAGGCTCCTCCCTCTCCCCTCCACTCTAATAGACTACCTAGGCGAATATCCGTACTCGATCTAA
Protein
MTLAARITPQNACLEVGVPLGGWSSGVACCPNCRHQLRVSLAPASGKCHALPSPHTPPFTVPPTYWPHSPLYAPPSPIVPAPYNDELKRLADTLSALRLSGWYYDTLNWQGARTLLKDASVGAFVVRDSGDRNFIFSLSVQTERGPTSVRLHYEQGFFRLDCDRPLARYMPRFRCVVEMVQQYRRAGQRGAAGTVWVDREGCPHSPVLLTHPLRKSPPSLMHAARLAVHKALDSNPLTPKLWCAPKHRLLPLPSTLIDYLGEYPYSI
Summary
Uniprot
H9J7U0
F2XHI7
A0A2A4JZA5
A0A2H1W606
A0A194QCF5
A0A194QTD5
+ More
A0A212EIU7 S4NSQ6 A0A0L7LJJ7 A0A1Y1KR34 V5I9A8 A0A2J7QEB9 A0A1W4WWN6 A0A2P8ZGQ1 A0A1B6JB10 A0A1B6LK57 A0A1B6FRU0 A0A1B6GJF9 A0A1B6LC24 A0A1B6EG95 A0A1B6FYR9 D6WVY8 L7MGA8 L7MGX2 A0A1Z5L4I6 A0A067QTM3 T1HD47 A0A0A9YZI6 A0A2R5LDF8 A0A023F0Z3 A0A1V9XP72 A0A224XVT4 D1G5I2 A0A0P4WFV5 E0VHG6 A0A2L2Y088 E2C1J5 A0A154PIK3 W4XHC4 A0A0P4VR82 E9IR72 A0A0K0YBD3 A0A310SA25 C3YM24 A0A3Q0IJH3 A0A026WLC2 A0A3L8DCU6
A0A212EIU7 S4NSQ6 A0A0L7LJJ7 A0A1Y1KR34 V5I9A8 A0A2J7QEB9 A0A1W4WWN6 A0A2P8ZGQ1 A0A1B6JB10 A0A1B6LK57 A0A1B6FRU0 A0A1B6GJF9 A0A1B6LC24 A0A1B6EG95 A0A1B6FYR9 D6WVY8 L7MGA8 L7MGX2 A0A1Z5L4I6 A0A067QTM3 T1HD47 A0A0A9YZI6 A0A2R5LDF8 A0A023F0Z3 A0A1V9XP72 A0A224XVT4 D1G5I2 A0A0P4WFV5 E0VHG6 A0A2L2Y088 E2C1J5 A0A154PIK3 W4XHC4 A0A0P4VR82 E9IR72 A0A0K0YBD3 A0A310SA25 C3YM24 A0A3Q0IJH3 A0A026WLC2 A0A3L8DCU6
Pubmed
EMBL
BABH01020142
HQ540306
ADZ99024.1
NWSH01000358
PCG77018.1
ODYU01006570
+ More
SOQ48521.1 KQ459232 KPJ02665.1 KQ461155 KPJ08265.1 AGBW02014579 OWR41390.1 GAIX01013972 JAA78588.1 JTDY01000867 KOB75622.1 GEZM01075900 JAV63889.1 GALX01003260 JAB65206.1 NEVH01015316 PNF26932.1 PYGN01000062 PSN55668.1 GECU01011315 JAS96391.1 GEBQ01015917 JAT24060.1 GECZ01030540 GECZ01017046 GECZ01015887 GECZ01014650 GECZ01010793 JAS39229.1 JAS52723.1 JAS53882.1 JAS55119.1 JAS58976.1 GECZ01030276 GECZ01026536 GECZ01017024 GECZ01013011 GECZ01007218 JAS39493.1 JAS43233.1 JAS52745.1 JAS56758.1 JAS62551.1 GEBQ01018749 JAT21228.1 GEDC01000387 JAS36911.1 GECZ01014447 JAS55322.1 KQ971359 EFA08217.1 GACK01001798 JAA63236.1 GACK01001799 JAA63235.1 GFJQ02004711 JAW02259.1 KK853286 KDR08949.1 ACPB03009573 GBHO01005162 GBHO01005161 GBHO01005160 GBRD01014823 GDHC01006644 GDHC01001396 JAG38442.1 JAG38443.1 JAG38444.1 JAG51003.1 JAQ11985.1 JAQ17233.1 GGLE01003372 MBY07498.1 GBBI01004026 JAC14686.1 MNPL01006790 OQR75138.1 GFTR01003856 JAW12570.1 FJ607955 ACU42699.1 GDRN01092046 GDRN01092045 JAI60164.1 DS235170 EEB12822.1 IAAA01006484 IAAA01006485 LAA01593.1 GL451937 EFN78171.1 KQ434924 KZC11642.1 AAGJ04061948 GDKW01003965 JAI52630.1 GL765099 EFZ16932.1 KR476664 AKS48289.1 KQ762541 OAD55668.1 GG666529 EEN58618.1 KK107167 EZA56456.1 QOIP01000010 RLU18126.1
SOQ48521.1 KQ459232 KPJ02665.1 KQ461155 KPJ08265.1 AGBW02014579 OWR41390.1 GAIX01013972 JAA78588.1 JTDY01000867 KOB75622.1 GEZM01075900 JAV63889.1 GALX01003260 JAB65206.1 NEVH01015316 PNF26932.1 PYGN01000062 PSN55668.1 GECU01011315 JAS96391.1 GEBQ01015917 JAT24060.1 GECZ01030540 GECZ01017046 GECZ01015887 GECZ01014650 GECZ01010793 JAS39229.1 JAS52723.1 JAS53882.1 JAS55119.1 JAS58976.1 GECZ01030276 GECZ01026536 GECZ01017024 GECZ01013011 GECZ01007218 JAS39493.1 JAS43233.1 JAS52745.1 JAS56758.1 JAS62551.1 GEBQ01018749 JAT21228.1 GEDC01000387 JAS36911.1 GECZ01014447 JAS55322.1 KQ971359 EFA08217.1 GACK01001798 JAA63236.1 GACK01001799 JAA63235.1 GFJQ02004711 JAW02259.1 KK853286 KDR08949.1 ACPB03009573 GBHO01005162 GBHO01005161 GBHO01005160 GBRD01014823 GDHC01006644 GDHC01001396 JAG38442.1 JAG38443.1 JAG38444.1 JAG51003.1 JAQ11985.1 JAQ17233.1 GGLE01003372 MBY07498.1 GBBI01004026 JAC14686.1 MNPL01006790 OQR75138.1 GFTR01003856 JAW12570.1 FJ607955 ACU42699.1 GDRN01092046 GDRN01092045 JAI60164.1 DS235170 EEB12822.1 IAAA01006484 IAAA01006485 LAA01593.1 GL451937 EFN78171.1 KQ434924 KZC11642.1 AAGJ04061948 GDKW01003965 JAI52630.1 GL765099 EFZ16932.1 KR476664 AKS48289.1 KQ762541 OAD55668.1 GG666529 EEN58618.1 KK107167 EZA56456.1 QOIP01000010 RLU18126.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9J7U0
F2XHI7
A0A2A4JZA5
A0A2H1W606
A0A194QCF5
A0A194QTD5
+ More
A0A212EIU7 S4NSQ6 A0A0L7LJJ7 A0A1Y1KR34 V5I9A8 A0A2J7QEB9 A0A1W4WWN6 A0A2P8ZGQ1 A0A1B6JB10 A0A1B6LK57 A0A1B6FRU0 A0A1B6GJF9 A0A1B6LC24 A0A1B6EG95 A0A1B6FYR9 D6WVY8 L7MGA8 L7MGX2 A0A1Z5L4I6 A0A067QTM3 T1HD47 A0A0A9YZI6 A0A2R5LDF8 A0A023F0Z3 A0A1V9XP72 A0A224XVT4 D1G5I2 A0A0P4WFV5 E0VHG6 A0A2L2Y088 E2C1J5 A0A154PIK3 W4XHC4 A0A0P4VR82 E9IR72 A0A0K0YBD3 A0A310SA25 C3YM24 A0A3Q0IJH3 A0A026WLC2 A0A3L8DCU6
A0A212EIU7 S4NSQ6 A0A0L7LJJ7 A0A1Y1KR34 V5I9A8 A0A2J7QEB9 A0A1W4WWN6 A0A2P8ZGQ1 A0A1B6JB10 A0A1B6LK57 A0A1B6FRU0 A0A1B6GJF9 A0A1B6LC24 A0A1B6EG95 A0A1B6FYR9 D6WVY8 L7MGA8 L7MGX2 A0A1Z5L4I6 A0A067QTM3 T1HD47 A0A0A9YZI6 A0A2R5LDF8 A0A023F0Z3 A0A1V9XP72 A0A224XVT4 D1G5I2 A0A0P4WFV5 E0VHG6 A0A2L2Y088 E2C1J5 A0A154PIK3 W4XHC4 A0A0P4VR82 E9IR72 A0A0K0YBD3 A0A310SA25 C3YM24 A0A3Q0IJH3 A0A026WLC2 A0A3L8DCU6
PDB
6I5J
E-value=1.81419e-21,
Score=251
Ontologies
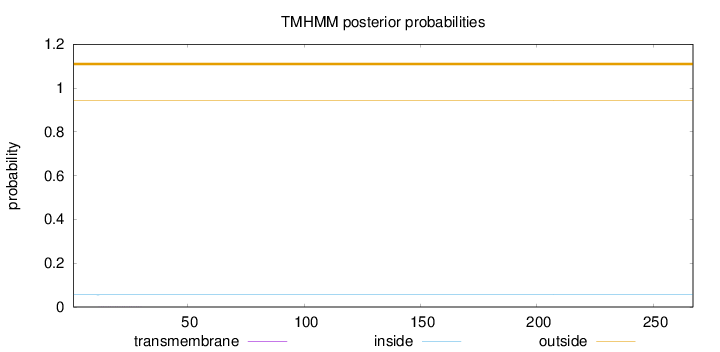
Topology
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00670999999999998
Exp number, first 60 AAs:
0.0063
Total prob of N-in:
0.05573
outside
1 - 267
Population Genetic Test Statistics
Pi
26.232041
Theta
26.41521
Tajima's D
-0.853254
CLR
0.019009
CSRT
0.165091745412729
Interpretation
Uncertain
