Pre Gene Modal
BGIBMGA005628
Annotation
uridine_5'-monophosphate_synthase_[Bombyx_mori]
Full name
Uridine 5'-monophosphate synthase
Alternative Name
Rudimentary-like protein
Location in the cell
Cytoplasmic Reliability : 1.974
Sequence
CDS
ATGGGTTTAGATAAACTAGAAGATCTAGCTTTAAAACTATTTTCTATTGATGCCGTGAAATTTGGAGATTTTACTACTAAGACCGGCATCAAAACACCGGCCTATTTTGATTTGAGGGTTATTGTTAGTTATCCCGACATCATGGAGCTTACCTCGAATATGTTGTACGATTTAGCCGTAAAAGACAGCCAATTTGATCATTTGTGTGGAGTTCCATATACTGCCTTGCCCATTGCTACACTACTAAGTATACAAGCGAAAAAACCTATGTTGATGAGAAGGAAAGAAACAAAATCTTATGGAACCAAAAAAAGCATTGAAGGTCATTTTAAAAAAGATGACACATGTCTCATCATCGAGGACGTCATAACATCTGGCTCAAGCATACTGGAAACAGTCAAAGATTTGAAAAATGAAGGGCTCGTAACTACAGAAGCTGCCATTATTTTAGACCGGGAGCAAGGTGGTAGGGAGAACTTAGCTAAAAATGGCATCCATGTAAAATCATTGTTCACAATGAGAACATTGATTGAAATACTTAAAAAACACAAGAAAATAACAGATGAAATTGTAGTTACAGTAAAAAATTACTTGAATGAAACAAAGGCTCCTGTTATAGAAAAACTTATGGAACGTGTTATACTACCATATGAAAAAAGAATAGATTTTGCAACAAATCTTGTAGCCAGGCAACTATTCAACATTATGGCCCCACAAAGAAAACTAATTTATGTCTCTCAGCAGACCTCACCATGA
Protein
MGLDKLEDLALKLFSIDAVKFGDFTTKTGIKTPAYFDLRVIVSYPDIMELTSNMLYDLAVKDSQFDHLCGVPYTALPIATLLSIQAKKPMLMRRKETKSYGTKKSIEGHFKKDDTCLIIEDVITSGSSILETVKDLKNEGLVTTEAAIILDREQGGRENLAKNGIHVKSLFTMRTLIEILKKHKKITDEIVVTVKNYLNETKAPVIEKLMERVILPYEKRIDFATNLVARQLFNIMAPQRKLIYVSQQTSP
Summary
Catalytic Activity
diphosphate + orotidine 5'-phosphate = 5-phospho-alpha-D-ribose 1-diphosphate + orotate
H(+) + orotidine 5'-phosphate = CO2 + UMP
H(+) + orotidine 5'-phosphate = CO2 + UMP
Similarity
In the N-terminal section; belongs to the purine/pyrimidine phosphoribosyltransferase family.
In the C-terminal section; belongs to the OMP decarboxylase family.
In the C-terminal section; belongs to the OMP decarboxylase family.
Keywords
Complete proteome
Decarboxylase
Glycosyltransferase
Lyase
Multifunctional enzyme
Pyrimidine biosynthesis
Reference proteome
Transferase
Feature
chain Uridine 5'-monophosphate synthase
Uniprot
H9J7Y6
Q2F642
A0A2A4JS88
A0A2H1V5H1
D0ABA9
A0A3S2M065
+ More
A0A0L7LET4 A0A194QGS7 A0A212EIU5 A0A194QXI0 S4NGX6 A0A0C9QBM4 A0A1L8DY49 K7IX88 A0A154PPC3 W5JU62 A0A2M4BL31 A0A1B0CG08 A0A2M4BL32 T1DQ66 A0A2M3Z341 A0A182SJA8 A0A2M4AG61 A0A023ES01 A0A0M8ZX99 A0A232FC27 T1GJV6 D1ZZL8 A0A182YGP3 A0A1B6K1V9 A0A1I8QBG3 B4JSX4 E2AMC4 A0A0K8TMZ9 A0A067RKE1 N6ST75 B0W331 A0A182QCX4 A0A1Q3F1P9 Q16QF3 A0A1S4FSU7 A0A182LTP8 A0A182IR66 A0A182HBI2 A0A1B6EQX9 A0A1B6G5N9 A0A340TBA6 A8HN92 E0VTC0 A0A2G8KIM2 A0A2A3E9E0 Q7QB57 B4R153 A0A182UWE0 B4IKL7 A0A182U319 Q01637 A0A336KMV5 B3P2Y9 A0A200PSJ9 A0A1W4VZ99 A0A0L0CLW6 C3ZXV8 A0A1B0A7K6 W4XIB1 Q293K2 A0A2P8Z2I2 A0A1A9YAD0 U5EU74 A0A182FSD4 A0A1L8ECV2 A0A182RYP4 A0A1B6KY40 R7U604 A0A3B0JMA6 B4NH48 A0A0B4J2P4 U5CZ43 B3M2W9 A0A1A9V6T5 A0A158NHC2 A0A2K3E7W9 B4GM06 A0A2I0K5N0 A0A026VYJ3 F4X083 A0A1B0FG80 A0A2I0X6A6 E9IMV3 B4M4V8 E9J1A5 A0A195D1G3 A0A182P6S0 S8DJP2 A0A0M3QXY6 A0A182NLC8 B4KE16 A0A2J7PLT5 A0A2J7PLS4 A0A182JTD1
A0A0L7LET4 A0A194QGS7 A0A212EIU5 A0A194QXI0 S4NGX6 A0A0C9QBM4 A0A1L8DY49 K7IX88 A0A154PPC3 W5JU62 A0A2M4BL31 A0A1B0CG08 A0A2M4BL32 T1DQ66 A0A2M3Z341 A0A182SJA8 A0A2M4AG61 A0A023ES01 A0A0M8ZX99 A0A232FC27 T1GJV6 D1ZZL8 A0A182YGP3 A0A1B6K1V9 A0A1I8QBG3 B4JSX4 E2AMC4 A0A0K8TMZ9 A0A067RKE1 N6ST75 B0W331 A0A182QCX4 A0A1Q3F1P9 Q16QF3 A0A1S4FSU7 A0A182LTP8 A0A182IR66 A0A182HBI2 A0A1B6EQX9 A0A1B6G5N9 A0A340TBA6 A8HN92 E0VTC0 A0A2G8KIM2 A0A2A3E9E0 Q7QB57 B4R153 A0A182UWE0 B4IKL7 A0A182U319 Q01637 A0A336KMV5 B3P2Y9 A0A200PSJ9 A0A1W4VZ99 A0A0L0CLW6 C3ZXV8 A0A1B0A7K6 W4XIB1 Q293K2 A0A2P8Z2I2 A0A1A9YAD0 U5EU74 A0A182FSD4 A0A1L8ECV2 A0A182RYP4 A0A1B6KY40 R7U604 A0A3B0JMA6 B4NH48 A0A0B4J2P4 U5CZ43 B3M2W9 A0A1A9V6T5 A0A158NHC2 A0A2K3E7W9 B4GM06 A0A2I0K5N0 A0A026VYJ3 F4X083 A0A1B0FG80 A0A2I0X6A6 E9IMV3 B4M4V8 E9J1A5 A0A195D1G3 A0A182P6S0 S8DJP2 A0A0M3QXY6 A0A182NLC8 B4KE16 A0A2J7PLT5 A0A2J7PLS4 A0A182JTD1
Pubmed
19121390
26227816
26354079
22118469
23622113
20075255
+ More
20920257 23761445 24945155 28648823 18362917 19820115 25244985 17994087 20798317 26369729 24845553 23537049 17510324 26483478 17932292 20566863 29023486 12364791 14747013 17210077 8444350 10731132 12537572 12537569 2122228 28552780 26108605 18563158 15632085 29403074 23254933 24357323 21347285 24508170 30249741 21719571 26754549 28902843 21282665 23855885
20920257 23761445 24945155 28648823 18362917 19820115 25244985 17994087 20798317 26369729 24845553 23537049 17510324 26483478 17932292 20566863 29023486 12364791 14747013 17210077 8444350 10731132 12537572 12537569 2122228 28552780 26108605 18563158 15632085 29403074 23254933 24357323 21347285 24508170 30249741 21719571 26754549 28902843 21282665 23855885
EMBL
BABH01020166
DQ311230
ABD36175.1
NWSH01000703
PCG74689.1
ODYU01000778
+ More
SOQ36083.1 FP102341 CBH09304.1 RSAL01000090 RVE48080.1 JTDY01001365 KOB74068.1 KQ459232 KPJ02651.1 AGBW02014579 OWR41391.1 KQ461155 KPJ08281.1 GAIX01014549 JAA78011.1 GBYB01011813 JAG81580.1 GFDF01002899 JAV11185.1 AAZX01000583 KQ435010 KZC13745.1 ADMH02000361 ETN66833.1 GGFJ01004639 MBW53780.1 AJWK01010631 GGFJ01004638 MBW53779.1 GAMD01002719 JAA98871.1 GGFM01002107 MBW22858.1 GGFK01006452 MBW39773.1 GAPW01001603 JAC11995.1 KQ435837 KOX71473.1 NNAY01000465 OXU28182.1 CAQQ02123440 KQ971338 EFA01834.2 GECU01002298 JAT05409.1 CH916373 EDV94864.1 GL440811 EFN65415.1 GDAI01002303 JAI15300.1 KK852421 KDR24287.1 APGK01057391 KB741280 KB631604 ENN70894.1 ERL84613.1 DS231830 EDS30739.1 AXCN02000121 GFDL01013586 JAV21459.1 CH477750 EAT36623.1 AXCM01003761 JXUM01032039 JXUM01032040 KQ560942 KXJ80255.1 GECZ01029408 JAS40361.1 GECZ01012037 JAS57732.1 DS496108 EDP09488.1 DS235761 EEB16626.1 MRZV01000554 PIK47842.1 KZ288313 PBC28347.1 AAAB01008880 EAA08525.3 CM000364 EDX12140.1 CH480853 EDW51621.1 L00968 AE014297 AY058714 X54230 UFQS01000645 UFQT01000410 UFQT01000645 SSX05697.1 SSX24054.1 CH954181 EDV48303.1 MVGT01004176 OVA01166.1 JRES01000211 KNC33256.1 GG666717 EEN42618.1 AAGJ04046045 AAGJ04046046 AAGJ04046047 CM000070 EAL29213.1 PYGN01000224 PSN50714.1 GANO01001613 JAB58258.1 GFDG01002333 JAV16466.1 GEBQ01023656 JAT16321.1 AMQN01001733 KB305005 ELU01511.1 OUUW01000007 SPP83397.1 CH964272 EDW84545.1 KI392503 ERN15444.1 CH902617 EDV43499.1 ADTU01015566 CM008962 PNW88880.1 CH479185 EDW38580.1 PGOL01000914 PKI63016.1 KK107566 QOIP01000006 EZA48873.1 RLU21950.1 GL888493 EGI60148.1 CCAG010021081 KZ502111 PKU83433.1 GL764255 EFZ18097.1 CH940652 EDW59669.1 GL767586 EFZ13419.1 KQ976973 KYN06707.1 AUSU01008136 EPS59712.1 CP012526 ALC46689.1 CH933806 EDW16037.1 NEVH01024423 PNF17279.1 PNF17277.1
SOQ36083.1 FP102341 CBH09304.1 RSAL01000090 RVE48080.1 JTDY01001365 KOB74068.1 KQ459232 KPJ02651.1 AGBW02014579 OWR41391.1 KQ461155 KPJ08281.1 GAIX01014549 JAA78011.1 GBYB01011813 JAG81580.1 GFDF01002899 JAV11185.1 AAZX01000583 KQ435010 KZC13745.1 ADMH02000361 ETN66833.1 GGFJ01004639 MBW53780.1 AJWK01010631 GGFJ01004638 MBW53779.1 GAMD01002719 JAA98871.1 GGFM01002107 MBW22858.1 GGFK01006452 MBW39773.1 GAPW01001603 JAC11995.1 KQ435837 KOX71473.1 NNAY01000465 OXU28182.1 CAQQ02123440 KQ971338 EFA01834.2 GECU01002298 JAT05409.1 CH916373 EDV94864.1 GL440811 EFN65415.1 GDAI01002303 JAI15300.1 KK852421 KDR24287.1 APGK01057391 KB741280 KB631604 ENN70894.1 ERL84613.1 DS231830 EDS30739.1 AXCN02000121 GFDL01013586 JAV21459.1 CH477750 EAT36623.1 AXCM01003761 JXUM01032039 JXUM01032040 KQ560942 KXJ80255.1 GECZ01029408 JAS40361.1 GECZ01012037 JAS57732.1 DS496108 EDP09488.1 DS235761 EEB16626.1 MRZV01000554 PIK47842.1 KZ288313 PBC28347.1 AAAB01008880 EAA08525.3 CM000364 EDX12140.1 CH480853 EDW51621.1 L00968 AE014297 AY058714 X54230 UFQS01000645 UFQT01000410 UFQT01000645 SSX05697.1 SSX24054.1 CH954181 EDV48303.1 MVGT01004176 OVA01166.1 JRES01000211 KNC33256.1 GG666717 EEN42618.1 AAGJ04046045 AAGJ04046046 AAGJ04046047 CM000070 EAL29213.1 PYGN01000224 PSN50714.1 GANO01001613 JAB58258.1 GFDG01002333 JAV16466.1 GEBQ01023656 JAT16321.1 AMQN01001733 KB305005 ELU01511.1 OUUW01000007 SPP83397.1 CH964272 EDW84545.1 KI392503 ERN15444.1 CH902617 EDV43499.1 ADTU01015566 CM008962 PNW88880.1 CH479185 EDW38580.1 PGOL01000914 PKI63016.1 KK107566 QOIP01000006 EZA48873.1 RLU21950.1 GL888493 EGI60148.1 CCAG010021081 KZ502111 PKU83433.1 GL764255 EFZ18097.1 CH940652 EDW59669.1 GL767586 EFZ13419.1 KQ976973 KYN06707.1 AUSU01008136 EPS59712.1 CP012526 ALC46689.1 CH933806 EDW16037.1 NEVH01024423 PNF17279.1 PNF17277.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000053268
UP000007151
+ More
UP000053240 UP000002358 UP000076502 UP000000673 UP000092461 UP000075901 UP000053105 UP000215335 UP000015102 UP000007266 UP000076408 UP000095300 UP000001070 UP000000311 UP000027135 UP000019118 UP000030742 UP000002320 UP000075886 UP000008820 UP000075883 UP000075880 UP000069940 UP000249989 UP000075920 UP000009046 UP000230750 UP000242457 UP000007062 UP000000304 UP000075903 UP000001292 UP000075902 UP000000803 UP000008711 UP000195402 UP000192221 UP000037069 UP000001554 UP000092445 UP000007110 UP000001819 UP000245037 UP000092443 UP000069272 UP000075900 UP000014760 UP000268350 UP000007798 UP000005203 UP000017836 UP000007801 UP000078200 UP000005205 UP000006906 UP000008744 UP000233551 UP000053097 UP000279307 UP000007755 UP000092444 UP000233837 UP000008792 UP000078542 UP000075885 UP000092553 UP000075884 UP000009192 UP000235965 UP000075881
UP000053240 UP000002358 UP000076502 UP000000673 UP000092461 UP000075901 UP000053105 UP000215335 UP000015102 UP000007266 UP000076408 UP000095300 UP000001070 UP000000311 UP000027135 UP000019118 UP000030742 UP000002320 UP000075886 UP000008820 UP000075883 UP000075880 UP000069940 UP000249989 UP000075920 UP000009046 UP000230750 UP000242457 UP000007062 UP000000304 UP000075903 UP000001292 UP000075902 UP000000803 UP000008711 UP000195402 UP000192221 UP000037069 UP000001554 UP000092445 UP000007110 UP000001819 UP000245037 UP000092443 UP000069272 UP000075900 UP000014760 UP000268350 UP000007798 UP000005203 UP000017836 UP000007801 UP000078200 UP000005205 UP000006906 UP000008744 UP000233551 UP000053097 UP000279307 UP000007755 UP000092444 UP000233837 UP000008792 UP000078542 UP000075885 UP000092553 UP000075884 UP000009192 UP000235965 UP000075881
Pfam
Interpro
IPR013785
Aldolase_TIM
+ More
IPR004467 Or_phspho_trans_dom
IPR023031 OPRT
IPR014732 OMPdecase
IPR011060 RibuloseP-bd_barrel
IPR000836 PRibTrfase_dom
IPR018089 OMPdecase_AS
IPR001754 OMPdeCOase_dom
IPR029057 PRTase-like
IPR036372 BEACH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR015507 rRNA-MeTfrase_E
IPR029063 SAM-dependent_MTases
IPR033523 IRE2
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR010513 KEN_dom
IPR018997 PUB_domain
IPR018391 PQQ_beta_propeller_repeat
IPR038357 KEN_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR004467 Or_phspho_trans_dom
IPR023031 OPRT
IPR014732 OMPdecase
IPR011060 RibuloseP-bd_barrel
IPR000836 PRibTrfase_dom
IPR018089 OMPdecase_AS
IPR001754 OMPdeCOase_dom
IPR029057 PRTase-like
IPR036372 BEACH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR015507 rRNA-MeTfrase_E
IPR029063 SAM-dependent_MTases
IPR033523 IRE2
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR010513 KEN_dom
IPR018997 PUB_domain
IPR018391 PQQ_beta_propeller_repeat
IPR038357 KEN_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
SUPFAM
ProteinModelPortal
H9J7Y6
Q2F642
A0A2A4JS88
A0A2H1V5H1
D0ABA9
A0A3S2M065
+ More
A0A0L7LET4 A0A194QGS7 A0A212EIU5 A0A194QXI0 S4NGX6 A0A0C9QBM4 A0A1L8DY49 K7IX88 A0A154PPC3 W5JU62 A0A2M4BL31 A0A1B0CG08 A0A2M4BL32 T1DQ66 A0A2M3Z341 A0A182SJA8 A0A2M4AG61 A0A023ES01 A0A0M8ZX99 A0A232FC27 T1GJV6 D1ZZL8 A0A182YGP3 A0A1B6K1V9 A0A1I8QBG3 B4JSX4 E2AMC4 A0A0K8TMZ9 A0A067RKE1 N6ST75 B0W331 A0A182QCX4 A0A1Q3F1P9 Q16QF3 A0A1S4FSU7 A0A182LTP8 A0A182IR66 A0A182HBI2 A0A1B6EQX9 A0A1B6G5N9 A0A340TBA6 A8HN92 E0VTC0 A0A2G8KIM2 A0A2A3E9E0 Q7QB57 B4R153 A0A182UWE0 B4IKL7 A0A182U319 Q01637 A0A336KMV5 B3P2Y9 A0A200PSJ9 A0A1W4VZ99 A0A0L0CLW6 C3ZXV8 A0A1B0A7K6 W4XIB1 Q293K2 A0A2P8Z2I2 A0A1A9YAD0 U5EU74 A0A182FSD4 A0A1L8ECV2 A0A182RYP4 A0A1B6KY40 R7U604 A0A3B0JMA6 B4NH48 A0A0B4J2P4 U5CZ43 B3M2W9 A0A1A9V6T5 A0A158NHC2 A0A2K3E7W9 B4GM06 A0A2I0K5N0 A0A026VYJ3 F4X083 A0A1B0FG80 A0A2I0X6A6 E9IMV3 B4M4V8 E9J1A5 A0A195D1G3 A0A182P6S0 S8DJP2 A0A0M3QXY6 A0A182NLC8 B4KE16 A0A2J7PLT5 A0A2J7PLS4 A0A182JTD1
A0A0L7LET4 A0A194QGS7 A0A212EIU5 A0A194QXI0 S4NGX6 A0A0C9QBM4 A0A1L8DY49 K7IX88 A0A154PPC3 W5JU62 A0A2M4BL31 A0A1B0CG08 A0A2M4BL32 T1DQ66 A0A2M3Z341 A0A182SJA8 A0A2M4AG61 A0A023ES01 A0A0M8ZX99 A0A232FC27 T1GJV6 D1ZZL8 A0A182YGP3 A0A1B6K1V9 A0A1I8QBG3 B4JSX4 E2AMC4 A0A0K8TMZ9 A0A067RKE1 N6ST75 B0W331 A0A182QCX4 A0A1Q3F1P9 Q16QF3 A0A1S4FSU7 A0A182LTP8 A0A182IR66 A0A182HBI2 A0A1B6EQX9 A0A1B6G5N9 A0A340TBA6 A8HN92 E0VTC0 A0A2G8KIM2 A0A2A3E9E0 Q7QB57 B4R153 A0A182UWE0 B4IKL7 A0A182U319 Q01637 A0A336KMV5 B3P2Y9 A0A200PSJ9 A0A1W4VZ99 A0A0L0CLW6 C3ZXV8 A0A1B0A7K6 W4XIB1 Q293K2 A0A2P8Z2I2 A0A1A9YAD0 U5EU74 A0A182FSD4 A0A1L8ECV2 A0A182RYP4 A0A1B6KY40 R7U604 A0A3B0JMA6 B4NH48 A0A0B4J2P4 U5CZ43 B3M2W9 A0A1A9V6T5 A0A158NHC2 A0A2K3E7W9 B4GM06 A0A2I0K5N0 A0A026VYJ3 F4X083 A0A1B0FG80 A0A2I0X6A6 E9IMV3 B4M4V8 E9J1A5 A0A195D1G3 A0A182P6S0 S8DJP2 A0A0M3QXY6 A0A182NLC8 B4KE16 A0A2J7PLT5 A0A2J7PLS4 A0A182JTD1
PDB
2WNS
E-value=5.21507e-47,
Score=471
Ontologies
PATHWAY
GO
PANTHER
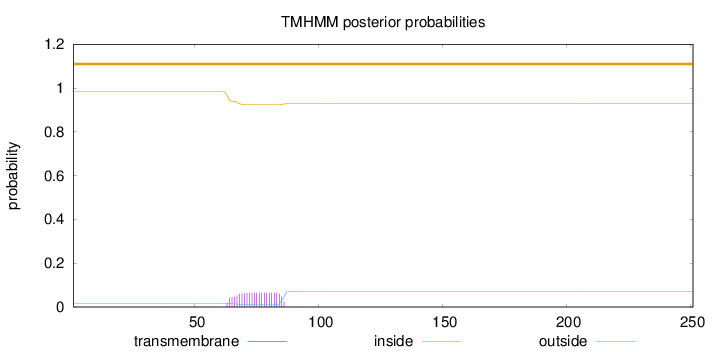
Topology
Length:
251
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.36901
Exp number, first 60 AAs:
0.0052
Total prob of N-in:
0.01597
outside
1 - 251
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
