Gene
KWMTBOMO16567
Annotation
PREDICTED:_pre-mRNA-splicing_factor_ATP-dependent_RNA_helicase_PRP16_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.337
Sequence
CDS
ATGCTACTGTTAAATGTAAAATTAATCTTGTTTATATGGGAAACTCCGTTTTTTTTTCTGTTCCAGGTGGTAGCTCGTCGCTCCGACCTGAAGTTGATCGTCACGTCAGCCACGATGGATTCGACTAAGTTTTCGACGTTTTTCGGTAACGTGCCGACTTTCACCATCCCCGGCAGGACCTTCCCCGTGGAGACGTTCTTCTCGAAGAACCCGTGCGAGGATTACGTTGACGGAGCTGTCAAACAGGCTTTACAAATTCATCTGCAGCCAGACGAGGGCGACATTCTGATATTCATGCCTGGTCAGGAAGACATCGAAGTTACGTGCGAGGTATTAACAGAAAGACTGGGCGATTTAGACAACGCGCCCCCCCTGACCGTCCTGCCGATATACTCTCAGCTCCCCGCTGACTTGCAGGCGAAGATATTCCAGCGAGCGCCGCCCGGACAGAGGAAGTGTATCGTCGCCACCAATATAGCAGAAACGTCACTAACCGTGGACGGGATCATGTACGTCATAGACTGCGGATATTGTAAGCTGAAGGTTTACAACCCGAGGATCGGTATGGACGCTTTGCAGATATATCCGGTTAGCCAGGCCAACGCCCGGCAGCGGGCGGGCCGAGCCGGGCGTACCGGCCCGGGCAAGGCCTTCTGTCTGTACACGCAGCGGCAGTACCAGCACGAGCTGCTGGCGTCTACTGTGCCGGAGATACAGAGGACGAACCTGGCGAACACCGTACTGTTGCTGAAGTCCTTGGGCGTTGAAGACCTACTGGCCTTCCACTTCATGGATCCCCCACCACAGGACACGATCCTGAACTCGATGTACCAGCTGTGGATCCTGGGCGCGCTGGACGGCACGGGCTCGCTGACGGCGCTCGGGCGGCAGATGGCGGAGTTCCCGCTGGACCCGGCGCAGTGCCACATGCTCATCGTGGCCGCGGACATGGGCTGCAGCGCCGAGGTGCTCATCATCGTGTCGATGCTGTCGGTGCCTTCAGTCTTCTACCGTCCTCAAGGTCGCGAGGAAGAAGCCGACGCAGTCAAAGAGAAATTCCAAGTCCCCGAATCCGATCACCTGACCCTCCTACACTTGTACAACCAGTGGAAATCAAACAACTACTCCAGTCAGTGGTGCACGGAGCACTTCGTTCACGCGAAGGCGATGAGGAAAGTGCGCGAGGTTCGCCAGCAGCTGAAGGACATTCTGCAACAGCAGCGGCTGCCGCTCGTGTCCTGCGGCACCGACTGGGACACCGTCAGGAAATGCATCTGTTCAGGTCCGTAG
Protein
MLLLNVKLILFIWETPFFFLFQVVARRSDLKLIVTSATMDSTKFSTFFGNVPTFTIPGRTFPVETFFSKNPCEDYVDGAVKQALQIHLQPDEGDILIFMPGQEDIEVTCEVLTERLGDLDNAPPLTVLPIYSQLPADLQAKIFQRAPPGQRKCIVATNIAETSLTVDGIMYVIDCGYCKLKVYNPRIGMDALQIYPVSQANARQRAGRAGRTGPGKAFCLYTQRQYQHELLASTVPEIQRTNLANTVLLLKSLGVEDLLAFHFMDPPPQDTILNSMYQLWILGALDGTGSLTALGRQMAEFPLDPAQCHMLIVAADMGCSAEVLIIVSMLSVPSVFYRPQGREEEADAVKEKFQVPESDHLTLLHLYNQWKSNNYSSQWCTEHFVHAKAMRKVREVRQQLKDILQQQRLPLVSCGTDWDTVRKCICSGP
Summary
Uniprot
A0A2A4K7G3
A0A212F809
A0A194QQ24
A0A2J7Q8P7
A0A1L8DH54
U4U7F1
+ More
A0A224XH31 A0A1W4X3L2 A0A1Y1NAW9 E0VLT3 A0A0A9VWS9 A0A182FLL1 W5JVK7 A0A182MD61 A0A182NB47 A0A182Y8H0 A0A182VTP7 Q7Q4Q1 A0A1W4VUB2 A0A182L037 A0A182IBU1 A0A182V576 A0A1S4GYI5 A0A182P4P2 B4JJA6 A0A182U2E2 A0A182X188 A0A182QLQ7 A0A1S4GZ85 B3NVV4 A0A182JED8 B4MEU2 A0A0K8VII5 A0A0A1X8I2 A0A182RLB0 B4Q2R1 A0A1B0D762 Q9VY54 A0A084W6V2 A0A1I8NLU6 A0A182GP01 A0A1I8MMG7 B4NBY3 Q29GN6 B4GVC8 B0W7R4 W8BTI0 B3N129 A0A1Q3F7K5 A0A0M4EP05 A0A3B0K5V8 A0A182SHR5 Q17KN2 N6TN26 A0A1I8NLU9 A0A336MAT9 A0A2S2QPY9 A0A1B6IRX8 A0A1J1INA1 A0A1A9XKI6 A0A1B0AFC4 A0A2P8YQD4 A0A2H8TP30 J9K217 A0A2S2PF50 L7MEK8 A0A0A1WZ92 A0A2A3E6K5 A0A369S956 A0A0J7L9M7 A0A1Z5KVH8 E9J412 A0A154P7H2 A0A195ERQ9 A0A2R5LAX9 F4WKH7 A0A195BQY3 A0A151XAZ2 A0A195DI32 K7IWW2 A0A158NQL4 A0A1Z5L0P5 A0A026WVY7 A0A139WKF7 E2BAH1 A0A195D3R1 A0A0C9RCK6 A0A087UM57 A0A293MHN9 A0A1E1X344 E1ZZK4 A0A2H1WTX0 A0A2P6KFB4 A0A0L0CFG7 A0A0P6HKN9 A0A164VWR5 A0A0P6G2Q7 V4B2P9 A0A232FCR5 A0A0P5W3D6
A0A224XH31 A0A1W4X3L2 A0A1Y1NAW9 E0VLT3 A0A0A9VWS9 A0A182FLL1 W5JVK7 A0A182MD61 A0A182NB47 A0A182Y8H0 A0A182VTP7 Q7Q4Q1 A0A1W4VUB2 A0A182L037 A0A182IBU1 A0A182V576 A0A1S4GYI5 A0A182P4P2 B4JJA6 A0A182U2E2 A0A182X188 A0A182QLQ7 A0A1S4GZ85 B3NVV4 A0A182JED8 B4MEU2 A0A0K8VII5 A0A0A1X8I2 A0A182RLB0 B4Q2R1 A0A1B0D762 Q9VY54 A0A084W6V2 A0A1I8NLU6 A0A182GP01 A0A1I8MMG7 B4NBY3 Q29GN6 B4GVC8 B0W7R4 W8BTI0 B3N129 A0A1Q3F7K5 A0A0M4EP05 A0A3B0K5V8 A0A182SHR5 Q17KN2 N6TN26 A0A1I8NLU9 A0A336MAT9 A0A2S2QPY9 A0A1B6IRX8 A0A1J1INA1 A0A1A9XKI6 A0A1B0AFC4 A0A2P8YQD4 A0A2H8TP30 J9K217 A0A2S2PF50 L7MEK8 A0A0A1WZ92 A0A2A3E6K5 A0A369S956 A0A0J7L9M7 A0A1Z5KVH8 E9J412 A0A154P7H2 A0A195ERQ9 A0A2R5LAX9 F4WKH7 A0A195BQY3 A0A151XAZ2 A0A195DI32 K7IWW2 A0A158NQL4 A0A1Z5L0P5 A0A026WVY7 A0A139WKF7 E2BAH1 A0A195D3R1 A0A0C9RCK6 A0A087UM57 A0A293MHN9 A0A1E1X344 E1ZZK4 A0A2H1WTX0 A0A2P6KFB4 A0A0L0CFG7 A0A0P6HKN9 A0A164VWR5 A0A0P6G2Q7 V4B2P9 A0A232FCR5 A0A0P5W3D6
Pubmed
22118469
26354079
23537049
28004739
20566863
25401762
+ More
26823975 20920257 23761445 25244985 12364791 20966253 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 26483478 25315136 15632085 24495485 17510324 29403074 25576852 30042472 28528879 21282665 21719571 20075255 21347285 24508170 30249741 18362917 19820115 20798317 28503490 26108605 23254933 28648823
26823975 20920257 23761445 25244985 12364791 20966253 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 26483478 25315136 15632085 24495485 17510324 29403074 25576852 30042472 28528879 21282665 21719571 20075255 21347285 24508170 30249741 18362917 19820115 20798317 28503490 26108605 23254933 28648823
EMBL
NWSH01000052
PCG80177.1
AGBW02009812
OWR49858.1
KQ461185
KPJ07464.1
+ More
NEVH01016949 PNF24954.1 GFDF01008367 JAV05717.1 KB631759 ERL85850.1 GFTR01008656 JAW07770.1 GEZM01007689 JAV94993.1 DS235281 EEB14339.1 GBHO01044941 GBRD01003505 GDHC01019435 JAF98662.1 JAG62316.1 JAP99193.1 ADMH02000297 ETN67065.1 AXCM01007720 AAAB01008964 EAA12175.4 APCN01000737 CH916370 EDV99658.1 AXCN02000643 CH954180 EDV47119.1 CH940664 EDW63067.1 GDHF01013637 JAI38677.1 GBXI01007314 JAD06978.1 CM000162 EDX01655.1 AJVK01026680 AE014298 AY058553 AAF48351.2 AAL13782.1 ATLV01020961 KE525310 KFB45946.1 JXUM01077340 KQ563003 KXJ74653.1 CH964239 EDW82342.1 CH379064 EAL32073.2 CH479192 EDW26415.1 DS231855 EDS38208.1 GAMC01009994 JAB96561.1 CH902650 EDV30064.1 GFDL01011536 JAV23509.1 CP012528 ALC49884.1 OUUW01000015 SPP88673.1 CH477222 EAT47272.1 APGK01058767 KB741291 ENN70610.1 UFQS01000316 UFQT01000316 SSX02787.1 SSX23158.1 GGMS01009989 MBY79192.1 GECU01018011 JAS89695.1 CVRI01000056 CRL01699.1 PYGN01000433 PSN46455.1 GFXV01003866 MBW15671.1 ABLF02035589 ABLF02035594 ABLF02035622 GGMR01015448 MBY28067.1 GACK01002767 JAA62267.1 GBXI01010301 JAD03991.1 KZ288349 PBC27393.1 NOWV01000044 RDD43091.1 LBMM01000146 KMR04745.1 GFJQ02007982 JAV98987.1 GL768069 EFZ12430.1 KQ434823 KZC07284.1 KQ982021 KYN30564.1 GGLE01002526 MBY06652.1 GL888199 EGI65315.1 KQ976419 KYM89293.1 KQ982335 KYQ57526.1 KQ980824 KYN12501.1 AAZX01007595 ADTU01023371 GFJQ02006008 GFJQ02004971 JAW00962.1 JAW01999.1 KK107087 QOIP01000001 EZA59856.1 RLU26921.1 KQ971330 KYB28331.1 GL446730 EFN87303.1 KQ976885 KYN07532.1 GBYB01010847 JAG80614.1 KK120525 KFM78446.1 GFWV01014910 MAA39639.1 GFAC01005732 JAT93456.1 GL435386 EFN73387.1 ODYU01011022 SOQ56501.1 MWRG01013534 PRD25022.1 JRES01000474 KNC30985.1 GDIQ01018775 JAN75962.1 LRGB01001361 KZS12737.1 GDIQ01041650 JAN53087.1 KB203854 ESO82809.1 NNAY01000459 OXU28239.1 GDIP01092526 JAM11189.1
NEVH01016949 PNF24954.1 GFDF01008367 JAV05717.1 KB631759 ERL85850.1 GFTR01008656 JAW07770.1 GEZM01007689 JAV94993.1 DS235281 EEB14339.1 GBHO01044941 GBRD01003505 GDHC01019435 JAF98662.1 JAG62316.1 JAP99193.1 ADMH02000297 ETN67065.1 AXCM01007720 AAAB01008964 EAA12175.4 APCN01000737 CH916370 EDV99658.1 AXCN02000643 CH954180 EDV47119.1 CH940664 EDW63067.1 GDHF01013637 JAI38677.1 GBXI01007314 JAD06978.1 CM000162 EDX01655.1 AJVK01026680 AE014298 AY058553 AAF48351.2 AAL13782.1 ATLV01020961 KE525310 KFB45946.1 JXUM01077340 KQ563003 KXJ74653.1 CH964239 EDW82342.1 CH379064 EAL32073.2 CH479192 EDW26415.1 DS231855 EDS38208.1 GAMC01009994 JAB96561.1 CH902650 EDV30064.1 GFDL01011536 JAV23509.1 CP012528 ALC49884.1 OUUW01000015 SPP88673.1 CH477222 EAT47272.1 APGK01058767 KB741291 ENN70610.1 UFQS01000316 UFQT01000316 SSX02787.1 SSX23158.1 GGMS01009989 MBY79192.1 GECU01018011 JAS89695.1 CVRI01000056 CRL01699.1 PYGN01000433 PSN46455.1 GFXV01003866 MBW15671.1 ABLF02035589 ABLF02035594 ABLF02035622 GGMR01015448 MBY28067.1 GACK01002767 JAA62267.1 GBXI01010301 JAD03991.1 KZ288349 PBC27393.1 NOWV01000044 RDD43091.1 LBMM01000146 KMR04745.1 GFJQ02007982 JAV98987.1 GL768069 EFZ12430.1 KQ434823 KZC07284.1 KQ982021 KYN30564.1 GGLE01002526 MBY06652.1 GL888199 EGI65315.1 KQ976419 KYM89293.1 KQ982335 KYQ57526.1 KQ980824 KYN12501.1 AAZX01007595 ADTU01023371 GFJQ02006008 GFJQ02004971 JAW00962.1 JAW01999.1 KK107087 QOIP01000001 EZA59856.1 RLU26921.1 KQ971330 KYB28331.1 GL446730 EFN87303.1 KQ976885 KYN07532.1 GBYB01010847 JAG80614.1 KK120525 KFM78446.1 GFWV01014910 MAA39639.1 GFAC01005732 JAT93456.1 GL435386 EFN73387.1 ODYU01011022 SOQ56501.1 MWRG01013534 PRD25022.1 JRES01000474 KNC30985.1 GDIQ01018775 JAN75962.1 LRGB01001361 KZS12737.1 GDIQ01041650 JAN53087.1 KB203854 ESO82809.1 NNAY01000459 OXU28239.1 GDIP01092526 JAM11189.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000235965
UP000030742
UP000192223
+ More
UP000009046 UP000069272 UP000000673 UP000075883 UP000075884 UP000076408 UP000075920 UP000007062 UP000192221 UP000075882 UP000075840 UP000075903 UP000075885 UP000001070 UP000075902 UP000076407 UP000075886 UP000008711 UP000075880 UP000008792 UP000075900 UP000002282 UP000092462 UP000000803 UP000030765 UP000095300 UP000069940 UP000249989 UP000095301 UP000007798 UP000001819 UP000008744 UP000002320 UP000007801 UP000092553 UP000268350 UP000075901 UP000008820 UP000019118 UP000183832 UP000092443 UP000092445 UP000245037 UP000007819 UP000242457 UP000253843 UP000036403 UP000076502 UP000078541 UP000007755 UP000078540 UP000075809 UP000078492 UP000002358 UP000005205 UP000053097 UP000279307 UP000007266 UP000008237 UP000078542 UP000054359 UP000000311 UP000037069 UP000076858 UP000030746 UP000215335
UP000009046 UP000069272 UP000000673 UP000075883 UP000075884 UP000076408 UP000075920 UP000007062 UP000192221 UP000075882 UP000075840 UP000075903 UP000075885 UP000001070 UP000075902 UP000076407 UP000075886 UP000008711 UP000075880 UP000008792 UP000075900 UP000002282 UP000092462 UP000000803 UP000030765 UP000095300 UP000069940 UP000249989 UP000095301 UP000007798 UP000001819 UP000008744 UP000002320 UP000007801 UP000092553 UP000268350 UP000075901 UP000008820 UP000019118 UP000183832 UP000092443 UP000092445 UP000245037 UP000007819 UP000242457 UP000253843 UP000036403 UP000076502 UP000078541 UP000007755 UP000078540 UP000075809 UP000078492 UP000002358 UP000005205 UP000053097 UP000279307 UP000007266 UP000008237 UP000078542 UP000054359 UP000000311 UP000037069 UP000076858 UP000030746 UP000215335
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2A4K7G3
A0A212F809
A0A194QQ24
A0A2J7Q8P7
A0A1L8DH54
U4U7F1
+ More
A0A224XH31 A0A1W4X3L2 A0A1Y1NAW9 E0VLT3 A0A0A9VWS9 A0A182FLL1 W5JVK7 A0A182MD61 A0A182NB47 A0A182Y8H0 A0A182VTP7 Q7Q4Q1 A0A1W4VUB2 A0A182L037 A0A182IBU1 A0A182V576 A0A1S4GYI5 A0A182P4P2 B4JJA6 A0A182U2E2 A0A182X188 A0A182QLQ7 A0A1S4GZ85 B3NVV4 A0A182JED8 B4MEU2 A0A0K8VII5 A0A0A1X8I2 A0A182RLB0 B4Q2R1 A0A1B0D762 Q9VY54 A0A084W6V2 A0A1I8NLU6 A0A182GP01 A0A1I8MMG7 B4NBY3 Q29GN6 B4GVC8 B0W7R4 W8BTI0 B3N129 A0A1Q3F7K5 A0A0M4EP05 A0A3B0K5V8 A0A182SHR5 Q17KN2 N6TN26 A0A1I8NLU9 A0A336MAT9 A0A2S2QPY9 A0A1B6IRX8 A0A1J1INA1 A0A1A9XKI6 A0A1B0AFC4 A0A2P8YQD4 A0A2H8TP30 J9K217 A0A2S2PF50 L7MEK8 A0A0A1WZ92 A0A2A3E6K5 A0A369S956 A0A0J7L9M7 A0A1Z5KVH8 E9J412 A0A154P7H2 A0A195ERQ9 A0A2R5LAX9 F4WKH7 A0A195BQY3 A0A151XAZ2 A0A195DI32 K7IWW2 A0A158NQL4 A0A1Z5L0P5 A0A026WVY7 A0A139WKF7 E2BAH1 A0A195D3R1 A0A0C9RCK6 A0A087UM57 A0A293MHN9 A0A1E1X344 E1ZZK4 A0A2H1WTX0 A0A2P6KFB4 A0A0L0CFG7 A0A0P6HKN9 A0A164VWR5 A0A0P6G2Q7 V4B2P9 A0A232FCR5 A0A0P5W3D6
A0A224XH31 A0A1W4X3L2 A0A1Y1NAW9 E0VLT3 A0A0A9VWS9 A0A182FLL1 W5JVK7 A0A182MD61 A0A182NB47 A0A182Y8H0 A0A182VTP7 Q7Q4Q1 A0A1W4VUB2 A0A182L037 A0A182IBU1 A0A182V576 A0A1S4GYI5 A0A182P4P2 B4JJA6 A0A182U2E2 A0A182X188 A0A182QLQ7 A0A1S4GZ85 B3NVV4 A0A182JED8 B4MEU2 A0A0K8VII5 A0A0A1X8I2 A0A182RLB0 B4Q2R1 A0A1B0D762 Q9VY54 A0A084W6V2 A0A1I8NLU6 A0A182GP01 A0A1I8MMG7 B4NBY3 Q29GN6 B4GVC8 B0W7R4 W8BTI0 B3N129 A0A1Q3F7K5 A0A0M4EP05 A0A3B0K5V8 A0A182SHR5 Q17KN2 N6TN26 A0A1I8NLU9 A0A336MAT9 A0A2S2QPY9 A0A1B6IRX8 A0A1J1INA1 A0A1A9XKI6 A0A1B0AFC4 A0A2P8YQD4 A0A2H8TP30 J9K217 A0A2S2PF50 L7MEK8 A0A0A1WZ92 A0A2A3E6K5 A0A369S956 A0A0J7L9M7 A0A1Z5KVH8 E9J412 A0A154P7H2 A0A195ERQ9 A0A2R5LAX9 F4WKH7 A0A195BQY3 A0A151XAZ2 A0A195DI32 K7IWW2 A0A158NQL4 A0A1Z5L0P5 A0A026WVY7 A0A139WKF7 E2BAH1 A0A195D3R1 A0A0C9RCK6 A0A087UM57 A0A293MHN9 A0A1E1X344 E1ZZK4 A0A2H1WTX0 A0A2P6KFB4 A0A0L0CFG7 A0A0P6HKN9 A0A164VWR5 A0A0P6G2Q7 V4B2P9 A0A232FCR5 A0A0P5W3D6
PDB
5YZG
E-value=0,
Score=1653
Ontologies
GO
Topology
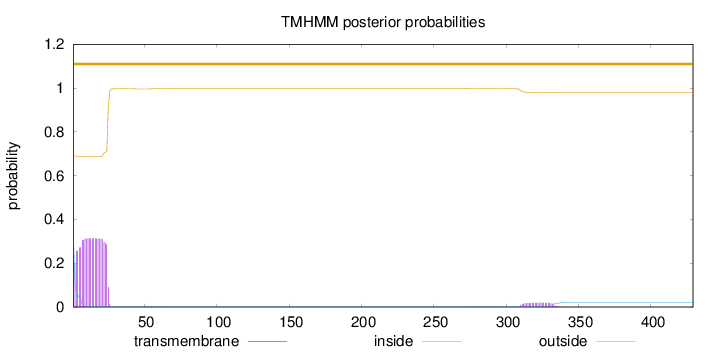
Length:
429
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.40448
Exp number, first 60 AAs:
6.92793
Total prob of N-in:
0.31085
outside
1 - 429
Population Genetic Test Statistics
Pi
19.781155
Theta
19.503407
Tajima's D
-1.498322
CLR
0.01655
CSRT
0.0602969851507425
Interpretation
Uncertain
