Gene
KWMTBOMO16565
Annotation
PREDICTED:_pre-mRNA-splicing_factor_ATP-dependent_RNA_helicase_PRP16-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.595 Mitochondrial Reliability : 1.852
Sequence
CDS
ATGACAAAGGCTTACTTCCAGCAGGCGGCGCGGCTGAAGGGCATCGGCGAGTACGTGAACTGCCGCACCGGCATGCCGTGCCACCTGCACCCCACCTCGGCGCTGTTCGGGCTGGGCTGCAGCCCCGACTACGTGGTGTACCACGAGCTCACCATGACGGCGCGCGAGTACATGCACTGCGTCACCGCCGTGGACGCGCGCTGGCTGGCCGAGCTGGGGCCCATGTTCTTCTCCGTCAAGGAGACCGGCAAATCGAATCGAGACAAGCGTAAAGAAGCAGCGGTCCACCTCCAACGCATGGAGGAAGAGATGAAGGAGGCCGAGGCCAAAATGGCAGAAGATCGCAAGAAGAAAGACGAAGACGTTCCCGTGAAGCAGGAGGTGGCCACCCCGGGGCTGGGCACCCCGCGCCGCACACCGCAGCGCCTGGGCCTGTAG
Protein
MTKAYFQQAARLKGIGEYVNCRTGMPCHLHPTSALFGLGCSPDYVVYHELTMTAREYMHCVTAVDARWLAELGPMFFSVKETGKSNRDKRKEAAVHLQRMEEEMKEAEAKMAEDRKKKDEDVPVKQEVATPGLGTPRRTPQRLGL
Summary
Uniprot
A0A194QQ24
A0A2H1WTX0
A0A2A4K7G3
A0A212F809
A0A194QFB1
A0A0L0CFG7
+ More
A0A0M4EP05 Q9VY50 A0A1L8DH54 B4NBY3 A0A1I8NLU8 A0A1I8MMG7 A0A1I8NLU6 T1P960 A0A1I8NLU9 B4JJA6 B4L3F7 Q9VY54 W8B600 B3NVV4 W8BTI0 B4R4N0 A0A0K8VII5 A0A0A1X8I2 A0A139WKF7 A0A0A1WZ92 B4MEU2 A0A1W4VUB2 B4Q2R1 W5JVK7 Q17KN2 B3N129 A0A182FLL1 A0A1Q3F7K5 A0A1W4X3L2 A0A1B0D762 A0A182SHR5 B0W7R4 A0A1A9VNH4 A0A1B0BTZ1 A0A1A9WID0 A0A1A9XKI6 A0A1B0AFC4 A0A1B6KNP3 A0A1B6C7F5 A0A182P4P2 A0A182Y8H0 A0A182MD61 A0A182JED8 A0A182VTP7 A0A182RLB0 A0A182U2E2 A0A182L037 A0A182NB47 A0A182V576 Q7Q4Q1 A0A1B6EWT2 A0A1S4GYI5 A0A1B6G162 A0A182IBU1 A0A084W6V2 A0A182QLQ7 A0A1S4GZ85 A0A182X188 A0A182GP01 A0A1J1INA1 N6TN26 A0A2J7Q8P7 U4U7F1 E0VLT3 A0A2P8YQD4 A0A1Y1NAW9 A0A088AJI9 A0A2A3E6K5 A0A0M8ZMK4 A0A0L7REB3 A0A195BQY3 A0A158NQL4 A0A151XAZ2 E9J412 A0A0J7L9M7 A0A195ERQ9 A0A195DI32 E1ZZK4 A0A195D3R1 F4WKH7 E2BAH1 A0A232FCR5 K7IWW2 A0A0A9VWS9 A0A026WVY7 A0A0C9RCK6 A0A1D2N344 T1I225 A0A131Z1J9 A0A224XH31 A0A2B4SVA6 A0A224YZH5
A0A0M4EP05 Q9VY50 A0A1L8DH54 B4NBY3 A0A1I8NLU8 A0A1I8MMG7 A0A1I8NLU6 T1P960 A0A1I8NLU9 B4JJA6 B4L3F7 Q9VY54 W8B600 B3NVV4 W8BTI0 B4R4N0 A0A0K8VII5 A0A0A1X8I2 A0A139WKF7 A0A0A1WZ92 B4MEU2 A0A1W4VUB2 B4Q2R1 W5JVK7 Q17KN2 B3N129 A0A182FLL1 A0A1Q3F7K5 A0A1W4X3L2 A0A1B0D762 A0A182SHR5 B0W7R4 A0A1A9VNH4 A0A1B0BTZ1 A0A1A9WID0 A0A1A9XKI6 A0A1B0AFC4 A0A1B6KNP3 A0A1B6C7F5 A0A182P4P2 A0A182Y8H0 A0A182MD61 A0A182JED8 A0A182VTP7 A0A182RLB0 A0A182U2E2 A0A182L037 A0A182NB47 A0A182V576 Q7Q4Q1 A0A1B6EWT2 A0A1S4GYI5 A0A1B6G162 A0A182IBU1 A0A084W6V2 A0A182QLQ7 A0A1S4GZ85 A0A182X188 A0A182GP01 A0A1J1INA1 N6TN26 A0A2J7Q8P7 U4U7F1 E0VLT3 A0A2P8YQD4 A0A1Y1NAW9 A0A088AJI9 A0A2A3E6K5 A0A0M8ZMK4 A0A0L7REB3 A0A195BQY3 A0A158NQL4 A0A151XAZ2 E9J412 A0A0J7L9M7 A0A195ERQ9 A0A195DI32 E1ZZK4 A0A195D3R1 F4WKH7 E2BAH1 A0A232FCR5 K7IWW2 A0A0A9VWS9 A0A026WVY7 A0A0C9RCK6 A0A1D2N344 T1I225 A0A131Z1J9 A0A224XH31 A0A2B4SVA6 A0A224YZH5
Pubmed
26354079
22118469
26108605
10731132
12537568
12537572
+ More
12537573 12537574 16110336 17569856 17569867 17994087 25315136 26109357 26109356 24495485 25830018 18362917 19820115 17550304 20920257 23761445 17510324 25244985 20966253 12364791 24438588 26483478 23537049 20566863 29403074 28004739 21347285 21282665 20798317 21719571 28648823 20075255 25401762 26823975 24508170 30249741 27289101 26830274 28797301
12537573 12537574 16110336 17569856 17569867 17994087 25315136 26109357 26109356 24495485 25830018 18362917 19820115 17550304 20920257 23761445 17510324 25244985 20966253 12364791 24438588 26483478 23537049 20566863 29403074 28004739 21347285 21282665 20798317 21719571 28648823 20075255 25401762 26823975 24508170 30249741 27289101 26830274 28797301
EMBL
KQ461185
KPJ07464.1
ODYU01011022
SOQ56501.1
NWSH01000052
PCG80177.1
+ More
AGBW02009812 OWR49858.1 KQ459252 KPJ02156.1 JRES01000474 KNC30985.1 CP012528 ALC49884.1 AE014298 AAF48355.2 GFDF01008367 JAV05717.1 CH964239 EDW82342.1 KA645252 AFP59881.1 CH916370 EDV99658.1 CH933810 EDW07085.2 AY058553 AAF48351.2 AAL13782.1 GAMC01009995 JAB96560.1 CH954180 EDV47119.1 GAMC01009994 JAB96561.1 CM000366 EDX17884.1 GDHF01013637 JAI38677.1 GBXI01007314 JAD06978.1 KQ971330 KYB28331.1 GBXI01010301 JAD03991.1 CH940664 EDW63067.1 CM000162 EDX01655.1 ADMH02000297 ETN67065.1 CH477222 EAT47272.1 CH902650 EDV30064.1 GFDL01011536 JAV23509.1 AJVK01026680 DS231855 EDS38208.1 JXJN01020416 GEBQ01027153 JAT12824.1 GEDC01027897 JAS09401.1 AXCM01007720 AAAB01008964 EAA12175.4 GECZ01027305 JAS42464.1 GECZ01013582 JAS56187.1 APCN01000737 ATLV01020961 KE525310 KFB45946.1 AXCN02000643 JXUM01077340 KQ563003 KXJ74653.1 CVRI01000056 CRL01699.1 APGK01058767 KB741291 ENN70610.1 NEVH01016949 PNF24954.1 KB631759 ERL85850.1 DS235281 EEB14339.1 PYGN01000433 PSN46455.1 GEZM01007689 JAV94993.1 KZ288349 PBC27393.1 KQ436240 KOX67360.1 KQ414612 KOC69165.1 KQ976419 KYM89293.1 ADTU01023371 KQ982335 KYQ57526.1 GL768069 EFZ12430.1 LBMM01000146 KMR04745.1 KQ982021 KYN30564.1 KQ980824 KYN12501.1 GL435386 EFN73387.1 KQ976885 KYN07532.1 GL888199 EGI65315.1 GL446730 EFN87303.1 NNAY01000459 OXU28239.1 AAZX01007595 GBHO01044941 GBRD01003505 GDHC01019435 JAF98662.1 JAG62316.1 JAP99193.1 KK107087 QOIP01000001 EZA59856.1 RLU26921.1 GBYB01010847 JAG80614.1 LJIJ01000279 ODM99474.1 ACPB03006723 GEDV01003942 JAP84615.1 GFTR01008656 JAW07770.1 LSMT01000015 PFX33109.1 GFPF01009963 MAA21109.1
AGBW02009812 OWR49858.1 KQ459252 KPJ02156.1 JRES01000474 KNC30985.1 CP012528 ALC49884.1 AE014298 AAF48355.2 GFDF01008367 JAV05717.1 CH964239 EDW82342.1 KA645252 AFP59881.1 CH916370 EDV99658.1 CH933810 EDW07085.2 AY058553 AAF48351.2 AAL13782.1 GAMC01009995 JAB96560.1 CH954180 EDV47119.1 GAMC01009994 JAB96561.1 CM000366 EDX17884.1 GDHF01013637 JAI38677.1 GBXI01007314 JAD06978.1 KQ971330 KYB28331.1 GBXI01010301 JAD03991.1 CH940664 EDW63067.1 CM000162 EDX01655.1 ADMH02000297 ETN67065.1 CH477222 EAT47272.1 CH902650 EDV30064.1 GFDL01011536 JAV23509.1 AJVK01026680 DS231855 EDS38208.1 JXJN01020416 GEBQ01027153 JAT12824.1 GEDC01027897 JAS09401.1 AXCM01007720 AAAB01008964 EAA12175.4 GECZ01027305 JAS42464.1 GECZ01013582 JAS56187.1 APCN01000737 ATLV01020961 KE525310 KFB45946.1 AXCN02000643 JXUM01077340 KQ563003 KXJ74653.1 CVRI01000056 CRL01699.1 APGK01058767 KB741291 ENN70610.1 NEVH01016949 PNF24954.1 KB631759 ERL85850.1 DS235281 EEB14339.1 PYGN01000433 PSN46455.1 GEZM01007689 JAV94993.1 KZ288349 PBC27393.1 KQ436240 KOX67360.1 KQ414612 KOC69165.1 KQ976419 KYM89293.1 ADTU01023371 KQ982335 KYQ57526.1 GL768069 EFZ12430.1 LBMM01000146 KMR04745.1 KQ982021 KYN30564.1 KQ980824 KYN12501.1 GL435386 EFN73387.1 KQ976885 KYN07532.1 GL888199 EGI65315.1 GL446730 EFN87303.1 NNAY01000459 OXU28239.1 AAZX01007595 GBHO01044941 GBRD01003505 GDHC01019435 JAF98662.1 JAG62316.1 JAP99193.1 KK107087 QOIP01000001 EZA59856.1 RLU26921.1 GBYB01010847 JAG80614.1 LJIJ01000279 ODM99474.1 ACPB03006723 GEDV01003942 JAP84615.1 GFTR01008656 JAW07770.1 LSMT01000015 PFX33109.1 GFPF01009963 MAA21109.1
Proteomes
UP000053240
UP000218220
UP000007151
UP000053268
UP000037069
UP000092553
+ More
UP000000803 UP000007798 UP000095300 UP000095301 UP000001070 UP000009192 UP000008711 UP000000304 UP000007266 UP000008792 UP000192221 UP000002282 UP000000673 UP000008820 UP000007801 UP000069272 UP000192223 UP000092462 UP000075901 UP000002320 UP000078200 UP000092460 UP000091820 UP000092443 UP000092445 UP000075885 UP000076408 UP000075883 UP000075880 UP000075920 UP000075900 UP000075902 UP000075882 UP000075884 UP000075903 UP000007062 UP000075840 UP000030765 UP000075886 UP000076407 UP000069940 UP000249989 UP000183832 UP000019118 UP000235965 UP000030742 UP000009046 UP000245037 UP000005203 UP000242457 UP000053105 UP000053825 UP000078540 UP000005205 UP000075809 UP000036403 UP000078541 UP000078492 UP000000311 UP000078542 UP000007755 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000094527 UP000015103 UP000225706
UP000000803 UP000007798 UP000095300 UP000095301 UP000001070 UP000009192 UP000008711 UP000000304 UP000007266 UP000008792 UP000192221 UP000002282 UP000000673 UP000008820 UP000007801 UP000069272 UP000192223 UP000092462 UP000075901 UP000002320 UP000078200 UP000092460 UP000091820 UP000092443 UP000092445 UP000075885 UP000076408 UP000075883 UP000075880 UP000075920 UP000075900 UP000075902 UP000075882 UP000075884 UP000075903 UP000007062 UP000075840 UP000030765 UP000075886 UP000076407 UP000069940 UP000249989 UP000183832 UP000019118 UP000235965 UP000030742 UP000009046 UP000245037 UP000005203 UP000242457 UP000053105 UP000053825 UP000078540 UP000005205 UP000075809 UP000036403 UP000078541 UP000078492 UP000000311 UP000078542 UP000007755 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000094527 UP000015103 UP000225706
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
A0A194QQ24
A0A2H1WTX0
A0A2A4K7G3
A0A212F809
A0A194QFB1
A0A0L0CFG7
+ More
A0A0M4EP05 Q9VY50 A0A1L8DH54 B4NBY3 A0A1I8NLU8 A0A1I8MMG7 A0A1I8NLU6 T1P960 A0A1I8NLU9 B4JJA6 B4L3F7 Q9VY54 W8B600 B3NVV4 W8BTI0 B4R4N0 A0A0K8VII5 A0A0A1X8I2 A0A139WKF7 A0A0A1WZ92 B4MEU2 A0A1W4VUB2 B4Q2R1 W5JVK7 Q17KN2 B3N129 A0A182FLL1 A0A1Q3F7K5 A0A1W4X3L2 A0A1B0D762 A0A182SHR5 B0W7R4 A0A1A9VNH4 A0A1B0BTZ1 A0A1A9WID0 A0A1A9XKI6 A0A1B0AFC4 A0A1B6KNP3 A0A1B6C7F5 A0A182P4P2 A0A182Y8H0 A0A182MD61 A0A182JED8 A0A182VTP7 A0A182RLB0 A0A182U2E2 A0A182L037 A0A182NB47 A0A182V576 Q7Q4Q1 A0A1B6EWT2 A0A1S4GYI5 A0A1B6G162 A0A182IBU1 A0A084W6V2 A0A182QLQ7 A0A1S4GZ85 A0A182X188 A0A182GP01 A0A1J1INA1 N6TN26 A0A2J7Q8P7 U4U7F1 E0VLT3 A0A2P8YQD4 A0A1Y1NAW9 A0A088AJI9 A0A2A3E6K5 A0A0M8ZMK4 A0A0L7REB3 A0A195BQY3 A0A158NQL4 A0A151XAZ2 E9J412 A0A0J7L9M7 A0A195ERQ9 A0A195DI32 E1ZZK4 A0A195D3R1 F4WKH7 E2BAH1 A0A232FCR5 K7IWW2 A0A0A9VWS9 A0A026WVY7 A0A0C9RCK6 A0A1D2N344 T1I225 A0A131Z1J9 A0A224XH31 A0A2B4SVA6 A0A224YZH5
A0A0M4EP05 Q9VY50 A0A1L8DH54 B4NBY3 A0A1I8NLU8 A0A1I8MMG7 A0A1I8NLU6 T1P960 A0A1I8NLU9 B4JJA6 B4L3F7 Q9VY54 W8B600 B3NVV4 W8BTI0 B4R4N0 A0A0K8VII5 A0A0A1X8I2 A0A139WKF7 A0A0A1WZ92 B4MEU2 A0A1W4VUB2 B4Q2R1 W5JVK7 Q17KN2 B3N129 A0A182FLL1 A0A1Q3F7K5 A0A1W4X3L2 A0A1B0D762 A0A182SHR5 B0W7R4 A0A1A9VNH4 A0A1B0BTZ1 A0A1A9WID0 A0A1A9XKI6 A0A1B0AFC4 A0A1B6KNP3 A0A1B6C7F5 A0A182P4P2 A0A182Y8H0 A0A182MD61 A0A182JED8 A0A182VTP7 A0A182RLB0 A0A182U2E2 A0A182L037 A0A182NB47 A0A182V576 Q7Q4Q1 A0A1B6EWT2 A0A1S4GYI5 A0A1B6G162 A0A182IBU1 A0A084W6V2 A0A182QLQ7 A0A1S4GZ85 A0A182X188 A0A182GP01 A0A1J1INA1 N6TN26 A0A2J7Q8P7 U4U7F1 E0VLT3 A0A2P8YQD4 A0A1Y1NAW9 A0A088AJI9 A0A2A3E6K5 A0A0M8ZMK4 A0A0L7REB3 A0A195BQY3 A0A158NQL4 A0A151XAZ2 E9J412 A0A0J7L9M7 A0A195ERQ9 A0A195DI32 E1ZZK4 A0A195D3R1 F4WKH7 E2BAH1 A0A232FCR5 K7IWW2 A0A0A9VWS9 A0A026WVY7 A0A0C9RCK6 A0A1D2N344 T1I225 A0A131Z1J9 A0A224XH31 A0A2B4SVA6 A0A224YZH5
PDB
5YZG
E-value=2.68981e-40,
Score=409
Ontologies
GO
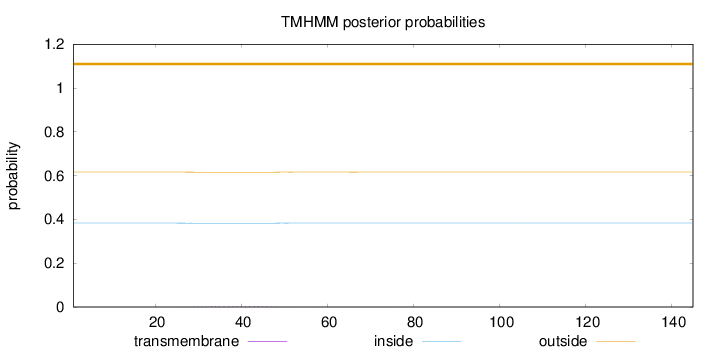
Topology
Length:
145
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05751
Exp number, first 60 AAs:
0.0529
Total prob of N-in:
0.38352
outside
1 - 145
Population Genetic Test Statistics
Pi
11.349496
Theta
15.221372
Tajima's D
-2.032501
CLR
0
CSRT
0.0112494375281236
Interpretation
Uncertain
