Pre Gene Modal
BGIBMGA012083
Annotation
PREDICTED:_ancient_ubiquitous_protein_1-like_[Bombyx_mori]
Full name
Ancient ubiquitous protein 1
Location in the cell
Mitochondrial Reliability : 1.853 PlasmaMembrane Reliability : 1.336
Sequence
CDS
ATGGTGATCAGAATATTCCTGGGTGTGATATTATGGATTGCATCAATTATATTGCCCAACAAATGGGCAGTCAGTCACATGTTAACTACTTTGGCTTGTTGGACTTTTGGTGTCTATGTGAAGCTCAAAGGACAAAGAGACCCAAGATGTAGTGTGTTGGTTGCTAACTATGTGACATGTTTCGATTCTCTTGCAGCTGCACATGTCTTTGGTACCATAAGTTTGAAAAGATGGAAGTTGCCACCATTTTTCGCATCGACGTTGGGTATAAAGAACGCCTCACAATTTTCTAAAAAGCAACATTTCTCTGAGGTGCCAACAAAGCCTGTGTTGATACAACCAGAAAGTGGACCCACGAACGGTAAAGCGGTGTTACGTTTCACAGATTGGCCATTCCAGATTAAAAGCAATGTGCAGCCGGTCGCCATCACCGTGGAGCGCGCCTTCACGCGTGCGACGGTGCAGCGGCGCCAGTACAGCGCGCGCTTCCCGTGGCGCGACGCGGCGTGGTTCCTGCTGTCGCCCGCCACCGTGTACACGCTGCACGCGCTGCCGCCGCTGCGCTGGGACGCGGGGGACGTCGGGGACCGCTGCCGGGCCGCCGTCGCAGAGGCGCTGCAGGTGGAAGTGTCGGAGCTGACGTGGGACGAGTTGTTCGCGGGGAGGCGCGTCCGGGCCGTCGCCGACCGCGCCGCCCCGCTCGTGCGCCTCGGGAACCAGGTCAAGGAAGTGTTGCCAGCCGTCCCACTCAAAACTATTCTCCAGGATTTAGAAAGAACTGGAAGCGTCGACATGACGATCACGAATTTCCTCGAGGGCGTGACTCCGTACGTCCCCGAAGATGCGGCCCCCGGACCGCAGCCCGGGCCCTCGGCGCCGCGCTACGTCGCGCCGCCGCCCACAGGAACCTTCCCGAAGACGGCCAAGGAACGACAGCTGAGCTTCGAGGAGCGAAAAGCTCAGATGATAGCGGAAGCTCGCAGGAGATATATCGAGAAACACGGACTGAACGTAACGTAG
Protein
MVIRIFLGVILWIASIILPNKWAVSHMLTTLACWTFGVYVKLKGQRDPRCSVLVANYVTCFDSLAAAHVFGTISLKRWKLPPFFASTLGIKNASQFSKKQHFSEVPTKPVLIQPESGPTNGKAVLRFTDWPFQIKSNVQPVAITVERAFTRATVQRRQYSARFPWRDAAWFLLSPATVYTLHALPPLRWDAGDVGDRCRAAVAEALQVEVSELTWDELFAGRRVRAVADRAAPLVRLGNQVKEVLPAVPLKTILQDLERTGSVDMTITNFLEGVTPYVPEDAAPGPQPGPSAPRYVAPPPTGTFPKTAKERQLSFEERKAQMIAEARRRYIEKHGLNVT
Summary
Description
May play a role in the translocation of terminally misfolded proteins from the endoplasmic reticulum lumen to the cytoplasm and their degradation by the proteasome.
Subunit
Identified in a complex that contains SEL1L, OS9, FAF2/UBXD8, UBE2J1/UBC6E and AUP1. Interacts with the cytoplasmic tail of ITGA2B, ITGA1, ITGA2, ITGA5, ITGAV and ITGAM.
Similarity
Belongs to the AUP1 family.
Keywords
Complete proteome
Endoplasmic reticulum
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Ubl conjugation pathway
3D-structure
Acetylation
Alternative splicing
Phosphoprotein
Feature
chain Ancient ubiquitous protein 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JRC3
A0A2A4K845
A0A212F821
A0A194QR67
A0A3S2M9U1
A0A194QAY6
+ More
D6WWW6 A0A2Z6FLD7 V5GFZ1 A0A1Y1N0D0 N6UAT9 A0A2J7QR91 A0A2J7QR90 E1ZXZ9 A0A023FAJ5 A0A224XQZ4 A0A0P4W545 R4FME3 A0A1W7RAY6 A0A232EKD5 A0A1B6G5A6 A0A069DU96 A0A0V0G4S0 A0A1B6KFF7 A0A0J7NCH3 A0A1S3H109 A0A2A3E6B4 A0A088AAK4 A0A0T6B5Y3 E2BUN6 A0A0A9VVG2 F4WCN5 M3Y9Q9 C3Y446 A0A151WY16 A0A195DFA4 A0A1S3GQ43 A0A195CVE3 A0A3B5BN49 A0A195ET26 A0A3Q3JQE0 K7JAV3 A0A158NHT8 A0A3B4YNB6 A0A3B4UHM2 V3ZNU5 H2T0T8 A0A195BVU4 A0A0L7QT10 A0A026W5G1 G5AXE7 A0A0P6J812 A0A0F8AFJ3 H0W5Z0 Q6PBN5 A0A3P9N8C1 A0A3Q1ES39 A0A3B3CBZ7 A0A3P8XIG9 A0A1A8G5G4 A0A1A8PKL4 A0A3B3XAY8 A0A1A8S2A2 A0A0P7WQI8 A0A2I4BDP7 A0A1W4ZMB1 A0A1A8B639 A0A146MZT2 A0A315VJZ4 A0A250YHG6 A0A1A8KZ50 A0A2U3VU15 H3AR98 A0A384CV96 A0A1A7Z196 A0A3Q7NS71 A0A3Q7V5L5 A0A1A8EGW0 F6XB90 R7U2I7 A0A2U3XV97 A0A2Y9HT56 W4ZDZ0 I3LXV8 M3W781 G1PU68 L9KS43 A0A2Y9JHL8 A0A341D3Z8 A0A384A9U6 A0A2F0B593 H0WHT3 G1T9C1 H2QI70 Q9Y679 A0A2Y9ETB8 A8KAQ6
D6WWW6 A0A2Z6FLD7 V5GFZ1 A0A1Y1N0D0 N6UAT9 A0A2J7QR91 A0A2J7QR90 E1ZXZ9 A0A023FAJ5 A0A224XQZ4 A0A0P4W545 R4FME3 A0A1W7RAY6 A0A232EKD5 A0A1B6G5A6 A0A069DU96 A0A0V0G4S0 A0A1B6KFF7 A0A0J7NCH3 A0A1S3H109 A0A2A3E6B4 A0A088AAK4 A0A0T6B5Y3 E2BUN6 A0A0A9VVG2 F4WCN5 M3Y9Q9 C3Y446 A0A151WY16 A0A195DFA4 A0A1S3GQ43 A0A195CVE3 A0A3B5BN49 A0A195ET26 A0A3Q3JQE0 K7JAV3 A0A158NHT8 A0A3B4YNB6 A0A3B4UHM2 V3ZNU5 H2T0T8 A0A195BVU4 A0A0L7QT10 A0A026W5G1 G5AXE7 A0A0P6J812 A0A0F8AFJ3 H0W5Z0 Q6PBN5 A0A3P9N8C1 A0A3Q1ES39 A0A3B3CBZ7 A0A3P8XIG9 A0A1A8G5G4 A0A1A8PKL4 A0A3B3XAY8 A0A1A8S2A2 A0A0P7WQI8 A0A2I4BDP7 A0A1W4ZMB1 A0A1A8B639 A0A146MZT2 A0A315VJZ4 A0A250YHG6 A0A1A8KZ50 A0A2U3VU15 H3AR98 A0A384CV96 A0A1A7Z196 A0A3Q7NS71 A0A3Q7V5L5 A0A1A8EGW0 F6XB90 R7U2I7 A0A2U3XV97 A0A2Y9HT56 W4ZDZ0 I3LXV8 M3W781 G1PU68 L9KS43 A0A2Y9JHL8 A0A341D3Z8 A0A384A9U6 A0A2F0B593 H0WHT3 G1T9C1 H2QI70 Q9Y679 A0A2Y9ETB8 A8KAQ6
Pubmed
19121390
22118469
26354079
18362917
19820115
28004739
+ More
23537049 20798317 25474469 27129103 28648823 26334808 25401762 26823975 21719571 18563158 20075255 21347285 23254933 21551351 24508170 30249741 21993625 25835551 21993624 29451363 25069045 29703783 28087693 9215903 16341006 17975172 23385571 16136131 11042152 14702039 15489334 12042322 18691976 18669648 18711132 19413330 20068231 21269460 22223895 22814378 23186163
23537049 20798317 25474469 27129103 28648823 26334808 25401762 26823975 21719571 18563158 20075255 21347285 23254933 21551351 24508170 30249741 21993625 25835551 21993624 29451363 25069045 29703783 28087693 9215903 16341006 17975172 23385571 16136131 11042152 14702039 15489334 12042322 18691976 18669648 18711132 19413330 20068231 21269460 22223895 22814378 23186163
EMBL
BABH01016087
BABH01016088
BABH01016089
NWSH01000052
PCG80176.1
AGBW02009812
+ More
OWR49859.1 KQ461185 KPJ07465.1 RSAL01000006 RVE54334.1 KQ459252 KPJ02155.1 KQ971361 EFA08762.1 LC384850 BBE28993.1 GALX01005567 JAB62899.1 GEZM01016147 JAV91362.1 APGK01042192 KB741002 KB631903 ENN75747.1 ERL87036.1 NEVH01011922 PNF31095.1 PNF31096.1 GL435158 EFN73939.1 GBBI01000774 JAC17938.1 GFTR01006027 JAW10399.1 GDKW01000097 JAI56498.1 ACPB03009974 GAHY01001043 JAA76467.1 GFAH01000076 JAV48313.1 NNAY01003812 OXU18819.1 GECZ01012276 JAS57493.1 GBGD01001612 JAC87277.1 GECL01003066 JAP03058.1 GEBQ01029806 JAT10171.1 LBMM01006854 KMQ90275.1 KZ288374 PBC26709.1 LJIG01009574 KRT82808.1 GL450732 EFN80589.1 GBHO01045401 GBRD01014303 GDHC01015658 JAF98202.1 JAG51523.1 JAQ02971.1 GL888070 EGI68215.1 AEYP01010579 GG666484 EEN65072.1 KQ982662 KYQ52647.1 KQ980905 KYN11522.1 KQ977259 KYN04605.1 KQ981979 KYN31311.1 AAZX01017979 ADTU01001748 KB201977 ESO93048.1 KQ976398 KYM92702.1 KQ414755 KOC61777.1 KK107403 QOIP01000011 EZA51312.1 RLU16719.1 JH167395 EHB01708.1 GEBF01003241 JAO00392.1 KQ042260 KKF16702.1 AAKN02036835 BC065447 BC059643 HAEB01019811 SBQ66338.1 HAEF01002135 HAEG01008467 SBR81945.1 HAEI01004260 HAEH01021019 SBS11683.1 JARO02007574 KPP63794.1 HADY01023465 HAEJ01010811 SBP61950.1 GCES01161149 JAQ25173.1 NHOQ01001578 PWA23482.1 GFFW01001711 JAV43077.1 HAED01001599 HAEE01016879 SBR36929.1 AFYH01154083 AFYH01154084 AFYH01154085 AFYH01154086 AFYH01154087 AFYH01154088 AFYH01154089 AFYH01154090 AFYH01154091 AFYH01154092 HADX01014430 SBP36662.1 HADZ01005287 HAEA01015844 SBQ44325.1 AAEX03010965 AMQN01009657 KB305899 ELU00555.1 AAGJ04129253 AGTP01057538 AANG04002548 AAPE02033107 KB320756 ELW63997.1 NTJE010074777 MBV98137.1 AAQR03101800 AAGW02006964 AACZ04063590 GABC01004058 GABF01009060 GABD01004021 GABE01005718 NBAG03000335 JAA07280.1 JAA13085.1 JAA29079.1 JAA39021.1 PNI38888.1 AF100754 AF100753 AF100746 AF165515 AK023983 CH471053 BC001658 BC033646 AK293121 BAF85810.1
OWR49859.1 KQ461185 KPJ07465.1 RSAL01000006 RVE54334.1 KQ459252 KPJ02155.1 KQ971361 EFA08762.1 LC384850 BBE28993.1 GALX01005567 JAB62899.1 GEZM01016147 JAV91362.1 APGK01042192 KB741002 KB631903 ENN75747.1 ERL87036.1 NEVH01011922 PNF31095.1 PNF31096.1 GL435158 EFN73939.1 GBBI01000774 JAC17938.1 GFTR01006027 JAW10399.1 GDKW01000097 JAI56498.1 ACPB03009974 GAHY01001043 JAA76467.1 GFAH01000076 JAV48313.1 NNAY01003812 OXU18819.1 GECZ01012276 JAS57493.1 GBGD01001612 JAC87277.1 GECL01003066 JAP03058.1 GEBQ01029806 JAT10171.1 LBMM01006854 KMQ90275.1 KZ288374 PBC26709.1 LJIG01009574 KRT82808.1 GL450732 EFN80589.1 GBHO01045401 GBRD01014303 GDHC01015658 JAF98202.1 JAG51523.1 JAQ02971.1 GL888070 EGI68215.1 AEYP01010579 GG666484 EEN65072.1 KQ982662 KYQ52647.1 KQ980905 KYN11522.1 KQ977259 KYN04605.1 KQ981979 KYN31311.1 AAZX01017979 ADTU01001748 KB201977 ESO93048.1 KQ976398 KYM92702.1 KQ414755 KOC61777.1 KK107403 QOIP01000011 EZA51312.1 RLU16719.1 JH167395 EHB01708.1 GEBF01003241 JAO00392.1 KQ042260 KKF16702.1 AAKN02036835 BC065447 BC059643 HAEB01019811 SBQ66338.1 HAEF01002135 HAEG01008467 SBR81945.1 HAEI01004260 HAEH01021019 SBS11683.1 JARO02007574 KPP63794.1 HADY01023465 HAEJ01010811 SBP61950.1 GCES01161149 JAQ25173.1 NHOQ01001578 PWA23482.1 GFFW01001711 JAV43077.1 HAED01001599 HAEE01016879 SBR36929.1 AFYH01154083 AFYH01154084 AFYH01154085 AFYH01154086 AFYH01154087 AFYH01154088 AFYH01154089 AFYH01154090 AFYH01154091 AFYH01154092 HADX01014430 SBP36662.1 HADZ01005287 HAEA01015844 SBQ44325.1 AAEX03010965 AMQN01009657 KB305899 ELU00555.1 AAGJ04129253 AGTP01057538 AANG04002548 AAPE02033107 KB320756 ELW63997.1 NTJE010074777 MBV98137.1 AAQR03101800 AAGW02006964 AACZ04063590 GABC01004058 GABF01009060 GABD01004021 GABE01005718 NBAG03000335 JAA07280.1 JAA13085.1 JAA29079.1 JAA39021.1 PNI38888.1 AF100754 AF100753 AF100746 AF165515 AK023983 CH471053 BC001658 BC033646 AK293121 BAF85810.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000283053
UP000053268
+ More
UP000007266 UP000019118 UP000030742 UP000235965 UP000000311 UP000015103 UP000215335 UP000036403 UP000085678 UP000242457 UP000005203 UP000008237 UP000007755 UP000000715 UP000001554 UP000075809 UP000078492 UP000081671 UP000078542 UP000261400 UP000078541 UP000261600 UP000002358 UP000005205 UP000261360 UP000261420 UP000030746 UP000005226 UP000078540 UP000053825 UP000053097 UP000279307 UP000006813 UP000005447 UP000000437 UP000242638 UP000257200 UP000261560 UP000265140 UP000261480 UP000034805 UP000192220 UP000192224 UP000265000 UP000245340 UP000008672 UP000261680 UP000286641 UP000286642 UP000002254 UP000014760 UP000245341 UP000248481 UP000007110 UP000005215 UP000011712 UP000001074 UP000011518 UP000248482 UP000252040 UP000261681 UP000005225 UP000001811 UP000002277 UP000005640 UP000248484
UP000007266 UP000019118 UP000030742 UP000235965 UP000000311 UP000015103 UP000215335 UP000036403 UP000085678 UP000242457 UP000005203 UP000008237 UP000007755 UP000000715 UP000001554 UP000075809 UP000078492 UP000081671 UP000078542 UP000261400 UP000078541 UP000261600 UP000002358 UP000005205 UP000261360 UP000261420 UP000030746 UP000005226 UP000078540 UP000053825 UP000053097 UP000279307 UP000006813 UP000005447 UP000000437 UP000242638 UP000257200 UP000261560 UP000265140 UP000261480 UP000034805 UP000192220 UP000192224 UP000265000 UP000245340 UP000008672 UP000261680 UP000286641 UP000286642 UP000002254 UP000014760 UP000245341 UP000248481 UP000007110 UP000005215 UP000011712 UP000001074 UP000011518 UP000248482 UP000252040 UP000261681 UP000005225 UP000001811 UP000002277 UP000005640 UP000248484
Pfam
PF02845 CUE
ProteinModelPortal
H9JRC3
A0A2A4K845
A0A212F821
A0A194QR67
A0A3S2M9U1
A0A194QAY6
+ More
D6WWW6 A0A2Z6FLD7 V5GFZ1 A0A1Y1N0D0 N6UAT9 A0A2J7QR91 A0A2J7QR90 E1ZXZ9 A0A023FAJ5 A0A224XQZ4 A0A0P4W545 R4FME3 A0A1W7RAY6 A0A232EKD5 A0A1B6G5A6 A0A069DU96 A0A0V0G4S0 A0A1B6KFF7 A0A0J7NCH3 A0A1S3H109 A0A2A3E6B4 A0A088AAK4 A0A0T6B5Y3 E2BUN6 A0A0A9VVG2 F4WCN5 M3Y9Q9 C3Y446 A0A151WY16 A0A195DFA4 A0A1S3GQ43 A0A195CVE3 A0A3B5BN49 A0A195ET26 A0A3Q3JQE0 K7JAV3 A0A158NHT8 A0A3B4YNB6 A0A3B4UHM2 V3ZNU5 H2T0T8 A0A195BVU4 A0A0L7QT10 A0A026W5G1 G5AXE7 A0A0P6J812 A0A0F8AFJ3 H0W5Z0 Q6PBN5 A0A3P9N8C1 A0A3Q1ES39 A0A3B3CBZ7 A0A3P8XIG9 A0A1A8G5G4 A0A1A8PKL4 A0A3B3XAY8 A0A1A8S2A2 A0A0P7WQI8 A0A2I4BDP7 A0A1W4ZMB1 A0A1A8B639 A0A146MZT2 A0A315VJZ4 A0A250YHG6 A0A1A8KZ50 A0A2U3VU15 H3AR98 A0A384CV96 A0A1A7Z196 A0A3Q7NS71 A0A3Q7V5L5 A0A1A8EGW0 F6XB90 R7U2I7 A0A2U3XV97 A0A2Y9HT56 W4ZDZ0 I3LXV8 M3W781 G1PU68 L9KS43 A0A2Y9JHL8 A0A341D3Z8 A0A384A9U6 A0A2F0B593 H0WHT3 G1T9C1 H2QI70 Q9Y679 A0A2Y9ETB8 A8KAQ6
D6WWW6 A0A2Z6FLD7 V5GFZ1 A0A1Y1N0D0 N6UAT9 A0A2J7QR91 A0A2J7QR90 E1ZXZ9 A0A023FAJ5 A0A224XQZ4 A0A0P4W545 R4FME3 A0A1W7RAY6 A0A232EKD5 A0A1B6G5A6 A0A069DU96 A0A0V0G4S0 A0A1B6KFF7 A0A0J7NCH3 A0A1S3H109 A0A2A3E6B4 A0A088AAK4 A0A0T6B5Y3 E2BUN6 A0A0A9VVG2 F4WCN5 M3Y9Q9 C3Y446 A0A151WY16 A0A195DFA4 A0A1S3GQ43 A0A195CVE3 A0A3B5BN49 A0A195ET26 A0A3Q3JQE0 K7JAV3 A0A158NHT8 A0A3B4YNB6 A0A3B4UHM2 V3ZNU5 H2T0T8 A0A195BVU4 A0A0L7QT10 A0A026W5G1 G5AXE7 A0A0P6J812 A0A0F8AFJ3 H0W5Z0 Q6PBN5 A0A3P9N8C1 A0A3Q1ES39 A0A3B3CBZ7 A0A3P8XIG9 A0A1A8G5G4 A0A1A8PKL4 A0A3B3XAY8 A0A1A8S2A2 A0A0P7WQI8 A0A2I4BDP7 A0A1W4ZMB1 A0A1A8B639 A0A146MZT2 A0A315VJZ4 A0A250YHG6 A0A1A8KZ50 A0A2U3VU15 H3AR98 A0A384CV96 A0A1A7Z196 A0A3Q7NS71 A0A3Q7V5L5 A0A1A8EGW0 F6XB90 R7U2I7 A0A2U3XV97 A0A2Y9HT56 W4ZDZ0 I3LXV8 M3W781 G1PU68 L9KS43 A0A2Y9JHL8 A0A341D3Z8 A0A384A9U6 A0A2F0B593 H0WHT3 G1T9C1 H2QI70 Q9Y679 A0A2Y9ETB8 A8KAQ6
PDB
2EKF
E-value=1.95073e-08,
Score=140
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
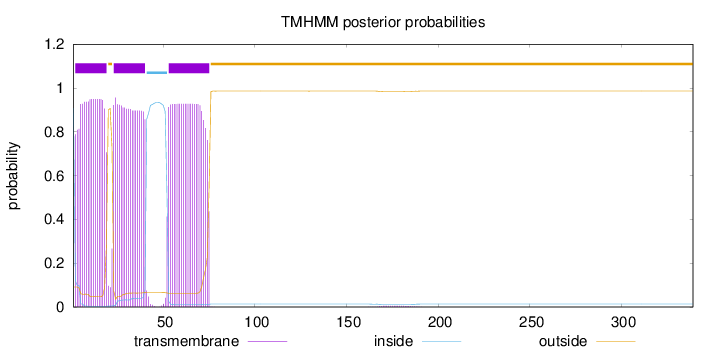
Length:
339
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
54.19903
Exp number, first 60 AAs:
41.0672
Total prob of N-in:
0.90839
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 22
TMhelix
23 - 40
inside
41 - 52
TMhelix
53 - 75
outside
76 - 339
Population Genetic Test Statistics
Pi
2.960212
Theta
24.572353
Tajima's D
-0.487858
CLR
0.136295
CSRT
0.251487425628719
Interpretation
Uncertain
