Gene
KWMTBOMO16563
Pre Gene Modal
BGIBMGA012072
Annotation
PREDICTED:_mucolipin-3-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.415 PlasmaMembrane Reliability : 1.597
Sequence
CDS
ATGGATGAAATCATGCATGCCCCTGATGATACTACCTGTTCTGAAGGCGAAGACGATCACAAAAAGGAAATTCGACCGAATGCCAACAATGAACAAGATTTTCCTCACCAATCGACTTCGTTCTCTCCGCCTACTCAAATGGAAGAAAAAATGCGTAGAAAACTGCAGTTTTTCTTTATGAATCCAATCGAGAAATGGAAAGCAAAGAGAAAATTTCCTTACAAATTTGTCGTGCAGGTAATCAAAATTGTACTTGTGACTTTTCAACTATGTTTATTCGCTCATAACAGATACAATCACGTGAACTATACATGGGATAACAGAATCACATTCTCGCATCTATTTTTACTCGGCTGGGACTCTACACGTGAGATAAACGCATATCCACCGGGAGCAGGACCACTTGCTTTGTACAAAATCGATGAGTTCTACAATACATTAGACTATGCAGTTGCCGGTTATACTAATTTATCTAATGCGATAGGCCCATATTCGTACAATGATGAAAATAACTTTATGCCAGATCCTGTCTTTTGCCAGTACAACTATAAACAAGGCATTATTTATGGCTTTAATGAGAGCTACCAGTTTAACTCTGAAGTCTTTGAGACCTGTATAAATTTCACAATTAAAGATAATGAAGATTTCAAATCTGAAGCGTACATTAGAGATGCTGGATTGAACATCAGTTTCTCATCATTAGTTAGAGCAAAATTAATGTTTTCCATCAAAACCATCAACTTCAGGGCTGCCGGTCCCATAACTCCACCAGACTGTTACCGGTTGATGTTGAAATAA
Protein
MDEIMHAPDDTTCSEGEDDHKKEIRPNANNEQDFPHQSTSFSPPTQMEEKMRRKLQFFFMNPIEKWKAKRKFPYKFVVQVIKIVLVTFQLCLFAHNRYNHVNYTWDNRITFSHLFLLGWDSTREINAYPPGAGPLALYKIDEFYNTLDYAVAGYTNLSNAIGPYSYNDENNFMPDPVFCQYNYKQGIIYGFNESYQFNSEVFETCINFTIKDNEDFKSEAYIRDAGLNISFSSLVRAKLMFSIKTINFRAAGPITPPDCYRLMLK
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
H9JRB2
A0A1E1WHP6
A0A2H1WSC3
A0A2A4K7C3
A0A0L7KYP6
A0A194QV84
+ More
A0A194Q9R9 S4PHF0 A0A2Z5U781 A0A212EQ52 A0A067R2P4 A0A2P8XTE4 A0A2J7R4H3 A0A2J7R4F7 A0A2J7R4F3 A0A2J7R4I6 A0A1W4WYY9 A0A195D3I4 D6WWW5 A0A182SYX8 A0A310SY03 A0A1Y1L746 A0A088ACW6 A0A1B6LQI0 A0A1B0C8F8 F4WWQ3 A0A182KS16 A0A1B6L400 T1DTU7 A0A2A3ETX7 A0A158NJ10 A0A1B6M9T8 A0A195BLC8 A0A1L8DDS5 A0A182XZB7 A0A182JTW3 A0A182QYK0 A0A2M4AD56 A0A2M4ADL2 A0A0L7QRA5 A0A2M4BGN8 W5JKF8 A0A182NHC0 A0A182X6L3 A7UR58 E1ZWV2 A0A2M3Z440 A0A2M3ZGP5 A0A151JNC3 A0A182U6M0 A0A026WKL4 A0A182RB16 A0A0J7KSA6 A0A023F518 A0A1B6DU29 A0A1B6DMH1 E2BH61 A0A1B6CB09 A0A151X792 A0A182WIY9 A0A154NX64 A0A182MPQ2 A0A384TJR2 W8AWP8 A0A1B6HEJ6 A0A1B6GAU7 T1I059 A0A1B6C196 A0A034VPR4 A0A0K8TXY5 A0A182FG26 A0A0A1WXU1 A0A2S2NXK7 A0A182GQQ2 A0A0C9R9Z5 A0A182G9W3 A0A182IX06 E9IIV9 B0WVU8 A0A1I8NRX0 J9JZ86 S6D9N2 Q17KW6 A0A1Q3FDF2 A0A1Q3FBR0 A0A2S2PY82 A0A1Q3FBT3 T1PBL3 A0A2H8TL45 A0A336KY26 A0A1B0C6N2 A0A1B0GF70 A0A1A9XJW7 A0A1A9ZDV9 A0A232EN81 K7IR03 A0A1A9UKP8 A0A0M4EQB7 A0A0L0C824 A0A1A9W5X5
A0A194Q9R9 S4PHF0 A0A2Z5U781 A0A212EQ52 A0A067R2P4 A0A2P8XTE4 A0A2J7R4H3 A0A2J7R4F7 A0A2J7R4F3 A0A2J7R4I6 A0A1W4WYY9 A0A195D3I4 D6WWW5 A0A182SYX8 A0A310SY03 A0A1Y1L746 A0A088ACW6 A0A1B6LQI0 A0A1B0C8F8 F4WWQ3 A0A182KS16 A0A1B6L400 T1DTU7 A0A2A3ETX7 A0A158NJ10 A0A1B6M9T8 A0A195BLC8 A0A1L8DDS5 A0A182XZB7 A0A182JTW3 A0A182QYK0 A0A2M4AD56 A0A2M4ADL2 A0A0L7QRA5 A0A2M4BGN8 W5JKF8 A0A182NHC0 A0A182X6L3 A7UR58 E1ZWV2 A0A2M3Z440 A0A2M3ZGP5 A0A151JNC3 A0A182U6M0 A0A026WKL4 A0A182RB16 A0A0J7KSA6 A0A023F518 A0A1B6DU29 A0A1B6DMH1 E2BH61 A0A1B6CB09 A0A151X792 A0A182WIY9 A0A154NX64 A0A182MPQ2 A0A384TJR2 W8AWP8 A0A1B6HEJ6 A0A1B6GAU7 T1I059 A0A1B6C196 A0A034VPR4 A0A0K8TXY5 A0A182FG26 A0A0A1WXU1 A0A2S2NXK7 A0A182GQQ2 A0A0C9R9Z5 A0A182G9W3 A0A182IX06 E9IIV9 B0WVU8 A0A1I8NRX0 J9JZ86 S6D9N2 Q17KW6 A0A1Q3FDF2 A0A1Q3FBR0 A0A2S2PY82 A0A1Q3FBT3 T1PBL3 A0A2H8TL45 A0A336KY26 A0A1B0C6N2 A0A1B0GF70 A0A1A9XJW7 A0A1A9ZDV9 A0A232EN81 K7IR03 A0A1A9UKP8 A0A0M4EQB7 A0A0L0C824 A0A1A9W5X5
Pubmed
19121390
26227816
26354079
23622113
22118469
24845553
+ More
29403074 18362917 19820115 28004739 21719571 20966253 21347285 25244985 20920257 23761445 12364791 14747013 17210077 20798317 24508170 30249741 25474469 24495485 25348373 25830018 26483478 21282665 23938757 17510324 25315136 28648823 20075255 26108605
29403074 18362917 19820115 28004739 21719571 20966253 21347285 25244985 20920257 23761445 12364791 14747013 17210077 20798317 24508170 30249741 25474469 24495485 25348373 25830018 26483478 21282665 23938757 17510324 25315136 28648823 20075255 26108605
EMBL
BABH01016087
GDQN01004566
JAT86488.1
ODYU01010693
SOQ55970.1
NWSH01000052
+ More
PCG80175.1 JTDY01004240 KOB68403.1 KQ461185 KPJ07466.1 KQ459252 KPJ02154.1 GAIX01000494 JAA92066.1 AP017502 BBB06793.1 AGBW02013309 OWR43618.1 KK852750 KDR17190.1 PYGN01001379 PSN35272.1 NEVH01007419 PNF35727.1 PNF35724.1 PNF35721.1 PNF35726.1 KQ976885 KYN07463.1 KQ971361 EFA09175.1 KQ759841 OAD62624.1 GEZM01064807 JAV68638.1 GEBQ01014019 JAT25958.1 AJWK01000543 GL888412 EGI61375.1 GEBQ01021505 JAT18472.1 GAMD01000744 JAB00847.1 KZ288185 PBC34954.1 ADTU01017350 ADTU01017351 GEBQ01007289 JAT32688.1 KQ976453 KYM85515.1 GFDF01009557 JAV04527.1 AXCN02000895 GGFK01005388 MBW38709.1 GGFK01005491 MBW38812.1 KQ414782 KOC61158.1 GGFJ01003085 MBW52226.1 ADMH02001258 ETN63365.1 AAAB01008807 EDO64504.1 GL434905 EFN74349.1 GGFM01002467 MBW23218.1 GGFM01006975 MBW27726.1 KQ978889 KYN27662.1 KK107163 QOIP01000002 EZA56523.1 RLU25816.1 LBMM01003811 KMQ93109.1 GBBI01002583 JAC16129.1 GEDC01008117 JAS29181.1 GEDC01010424 JAS26874.1 GL448268 EFN84982.1 GEDC01030678 GEDC01026620 GEDC01005880 GEDC01002170 JAS06620.1 JAS10678.1 JAS31418.1 JAS35128.1 KQ982454 KYQ56180.1 KQ434777 KZC04152.1 AXCM01000589 KX249700 AOR81477.1 GAMC01016048 JAB90507.1 GECU01034559 JAS73147.1 GECZ01010205 JAS59564.1 ACPB03011941 GEDC01030005 JAS07293.1 GAKP01015399 GAKP01015397 JAC43553.1 GDHF01032990 GDHF01027057 JAI19324.1 JAI25257.1 GBXI01010418 JAD03874.1 GGMR01009352 MBY21971.1 JXUM01015881 KQ560442 KXJ82386.1 GBYB01006334 GBYB01013234 JAG76101.1 JAG83001.1 JXUM01049861 KQ561624 KXJ77966.1 GL763562 EFZ19447.1 DS232132 EDS35780.1 ABLF02022564 HF586479 CCQ71383.1 CH477221 EAT47288.1 GFDL01009472 JAV25573.1 GFDL01010031 JAV25014.1 GGMS01001274 MBY70477.1 GFDL01010000 JAV25045.1 KA646156 AFP60785.1 GFXV01003061 MBW14866.1 UFQS01001196 UFQT01001196 SSX09641.1 SSX29437.1 JXJN01026843 CCAG010012768 NNAY01003212 OXU19782.1 CP012525 ALC44442.1 JRES01000760 KNC28588.1
PCG80175.1 JTDY01004240 KOB68403.1 KQ461185 KPJ07466.1 KQ459252 KPJ02154.1 GAIX01000494 JAA92066.1 AP017502 BBB06793.1 AGBW02013309 OWR43618.1 KK852750 KDR17190.1 PYGN01001379 PSN35272.1 NEVH01007419 PNF35727.1 PNF35724.1 PNF35721.1 PNF35726.1 KQ976885 KYN07463.1 KQ971361 EFA09175.1 KQ759841 OAD62624.1 GEZM01064807 JAV68638.1 GEBQ01014019 JAT25958.1 AJWK01000543 GL888412 EGI61375.1 GEBQ01021505 JAT18472.1 GAMD01000744 JAB00847.1 KZ288185 PBC34954.1 ADTU01017350 ADTU01017351 GEBQ01007289 JAT32688.1 KQ976453 KYM85515.1 GFDF01009557 JAV04527.1 AXCN02000895 GGFK01005388 MBW38709.1 GGFK01005491 MBW38812.1 KQ414782 KOC61158.1 GGFJ01003085 MBW52226.1 ADMH02001258 ETN63365.1 AAAB01008807 EDO64504.1 GL434905 EFN74349.1 GGFM01002467 MBW23218.1 GGFM01006975 MBW27726.1 KQ978889 KYN27662.1 KK107163 QOIP01000002 EZA56523.1 RLU25816.1 LBMM01003811 KMQ93109.1 GBBI01002583 JAC16129.1 GEDC01008117 JAS29181.1 GEDC01010424 JAS26874.1 GL448268 EFN84982.1 GEDC01030678 GEDC01026620 GEDC01005880 GEDC01002170 JAS06620.1 JAS10678.1 JAS31418.1 JAS35128.1 KQ982454 KYQ56180.1 KQ434777 KZC04152.1 AXCM01000589 KX249700 AOR81477.1 GAMC01016048 JAB90507.1 GECU01034559 JAS73147.1 GECZ01010205 JAS59564.1 ACPB03011941 GEDC01030005 JAS07293.1 GAKP01015399 GAKP01015397 JAC43553.1 GDHF01032990 GDHF01027057 JAI19324.1 JAI25257.1 GBXI01010418 JAD03874.1 GGMR01009352 MBY21971.1 JXUM01015881 KQ560442 KXJ82386.1 GBYB01006334 GBYB01013234 JAG76101.1 JAG83001.1 JXUM01049861 KQ561624 KXJ77966.1 GL763562 EFZ19447.1 DS232132 EDS35780.1 ABLF02022564 HF586479 CCQ71383.1 CH477221 EAT47288.1 GFDL01009472 JAV25573.1 GFDL01010031 JAV25014.1 GGMS01001274 MBY70477.1 GFDL01010000 JAV25045.1 KA646156 AFP60785.1 GFXV01003061 MBW14866.1 UFQS01001196 UFQT01001196 SSX09641.1 SSX29437.1 JXJN01026843 CCAG010012768 NNAY01003212 OXU19782.1 CP012525 ALC44442.1 JRES01000760 KNC28588.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000245037 UP000235965 UP000192223 UP000078542 UP000007266 UP000075901 UP000005203 UP000092461 UP000007755 UP000075882 UP000242457 UP000005205 UP000078540 UP000076408 UP000075881 UP000075886 UP000053825 UP000000673 UP000075884 UP000076407 UP000007062 UP000000311 UP000078492 UP000075902 UP000053097 UP000279307 UP000075900 UP000036403 UP000008237 UP000075809 UP000075920 UP000076502 UP000075883 UP000015103 UP000069272 UP000069940 UP000249989 UP000075880 UP000002320 UP000095300 UP000007819 UP000008820 UP000095301 UP000092460 UP000092444 UP000092443 UP000092445 UP000215335 UP000002358 UP000078200 UP000092553 UP000037069 UP000091820
UP000027135 UP000245037 UP000235965 UP000192223 UP000078542 UP000007266 UP000075901 UP000005203 UP000092461 UP000007755 UP000075882 UP000242457 UP000005205 UP000078540 UP000076408 UP000075881 UP000075886 UP000053825 UP000000673 UP000075884 UP000076407 UP000007062 UP000000311 UP000078492 UP000075902 UP000053097 UP000279307 UP000075900 UP000036403 UP000008237 UP000075809 UP000075920 UP000076502 UP000075883 UP000015103 UP000069272 UP000069940 UP000249989 UP000075880 UP000002320 UP000095300 UP000007819 UP000008820 UP000095301 UP000092460 UP000092444 UP000092443 UP000092445 UP000215335 UP000002358 UP000078200 UP000092553 UP000037069 UP000091820
PRIDE
Pfam
Interpro
IPR013122
PKD1_2_channel
+ More
IPR039031 Mucolipin
IPR035965 PAS-like_dom_sf
IPR000014 PAS
IPR027359 Volt_channel_dom_sf
IPR006029 Neurotrans-gated_channel_TM
IPR029000 Cyclophilin-like_dom_sf
IPR030172 KCNH6
IPR020892 Cyclophilin-type_PPIase_CS
IPR000595 cNMP-bd_dom
IPR006202 Neur_chan_lig-bd
IPR038105 Kif23_Arf-bd_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR036734 Neur_chan_lig-bd_sf
IPR032384 Kif23_Arf-bd
IPR005821 Ion_trans_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR027417 P-loop_NTPase
IPR014710 RmlC-like_jellyroll
IPR019821 Kinesin_motor_CS
IPR018490 cNMP-bd-like
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR003967 K_chnl_volt-dep_ERG
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR039031 Mucolipin
IPR035965 PAS-like_dom_sf
IPR000014 PAS
IPR027359 Volt_channel_dom_sf
IPR006029 Neurotrans-gated_channel_TM
IPR029000 Cyclophilin-like_dom_sf
IPR030172 KCNH6
IPR020892 Cyclophilin-type_PPIase_CS
IPR000595 cNMP-bd_dom
IPR006202 Neur_chan_lig-bd
IPR038105 Kif23_Arf-bd_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR036734 Neur_chan_lig-bd_sf
IPR032384 Kif23_Arf-bd
IPR005821 Ion_trans_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR027417 P-loop_NTPase
IPR014710 RmlC-like_jellyroll
IPR019821 Kinesin_motor_CS
IPR018490 cNMP-bd-like
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR003967 K_chnl_volt-dep_ERG
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
SUPFAM
ProteinModelPortal
H9JRB2
A0A1E1WHP6
A0A2H1WSC3
A0A2A4K7C3
A0A0L7KYP6
A0A194QV84
+ More
A0A194Q9R9 S4PHF0 A0A2Z5U781 A0A212EQ52 A0A067R2P4 A0A2P8XTE4 A0A2J7R4H3 A0A2J7R4F7 A0A2J7R4F3 A0A2J7R4I6 A0A1W4WYY9 A0A195D3I4 D6WWW5 A0A182SYX8 A0A310SY03 A0A1Y1L746 A0A088ACW6 A0A1B6LQI0 A0A1B0C8F8 F4WWQ3 A0A182KS16 A0A1B6L400 T1DTU7 A0A2A3ETX7 A0A158NJ10 A0A1B6M9T8 A0A195BLC8 A0A1L8DDS5 A0A182XZB7 A0A182JTW3 A0A182QYK0 A0A2M4AD56 A0A2M4ADL2 A0A0L7QRA5 A0A2M4BGN8 W5JKF8 A0A182NHC0 A0A182X6L3 A7UR58 E1ZWV2 A0A2M3Z440 A0A2M3ZGP5 A0A151JNC3 A0A182U6M0 A0A026WKL4 A0A182RB16 A0A0J7KSA6 A0A023F518 A0A1B6DU29 A0A1B6DMH1 E2BH61 A0A1B6CB09 A0A151X792 A0A182WIY9 A0A154NX64 A0A182MPQ2 A0A384TJR2 W8AWP8 A0A1B6HEJ6 A0A1B6GAU7 T1I059 A0A1B6C196 A0A034VPR4 A0A0K8TXY5 A0A182FG26 A0A0A1WXU1 A0A2S2NXK7 A0A182GQQ2 A0A0C9R9Z5 A0A182G9W3 A0A182IX06 E9IIV9 B0WVU8 A0A1I8NRX0 J9JZ86 S6D9N2 Q17KW6 A0A1Q3FDF2 A0A1Q3FBR0 A0A2S2PY82 A0A1Q3FBT3 T1PBL3 A0A2H8TL45 A0A336KY26 A0A1B0C6N2 A0A1B0GF70 A0A1A9XJW7 A0A1A9ZDV9 A0A232EN81 K7IR03 A0A1A9UKP8 A0A0M4EQB7 A0A0L0C824 A0A1A9W5X5
A0A194Q9R9 S4PHF0 A0A2Z5U781 A0A212EQ52 A0A067R2P4 A0A2P8XTE4 A0A2J7R4H3 A0A2J7R4F7 A0A2J7R4F3 A0A2J7R4I6 A0A1W4WYY9 A0A195D3I4 D6WWW5 A0A182SYX8 A0A310SY03 A0A1Y1L746 A0A088ACW6 A0A1B6LQI0 A0A1B0C8F8 F4WWQ3 A0A182KS16 A0A1B6L400 T1DTU7 A0A2A3ETX7 A0A158NJ10 A0A1B6M9T8 A0A195BLC8 A0A1L8DDS5 A0A182XZB7 A0A182JTW3 A0A182QYK0 A0A2M4AD56 A0A2M4ADL2 A0A0L7QRA5 A0A2M4BGN8 W5JKF8 A0A182NHC0 A0A182X6L3 A7UR58 E1ZWV2 A0A2M3Z440 A0A2M3ZGP5 A0A151JNC3 A0A182U6M0 A0A026WKL4 A0A182RB16 A0A0J7KSA6 A0A023F518 A0A1B6DU29 A0A1B6DMH1 E2BH61 A0A1B6CB09 A0A151X792 A0A182WIY9 A0A154NX64 A0A182MPQ2 A0A384TJR2 W8AWP8 A0A1B6HEJ6 A0A1B6GAU7 T1I059 A0A1B6C196 A0A034VPR4 A0A0K8TXY5 A0A182FG26 A0A0A1WXU1 A0A2S2NXK7 A0A182GQQ2 A0A0C9R9Z5 A0A182G9W3 A0A182IX06 E9IIV9 B0WVU8 A0A1I8NRX0 J9JZ86 S6D9N2 Q17KW6 A0A1Q3FDF2 A0A1Q3FBR0 A0A2S2PY82 A0A1Q3FBT3 T1PBL3 A0A2H8TL45 A0A336KY26 A0A1B0C6N2 A0A1B0GF70 A0A1A9XJW7 A0A1A9ZDV9 A0A232EN81 K7IR03 A0A1A9UKP8 A0A0M4EQB7 A0A0L0C824 A0A1A9W5X5
PDB
6AYG
E-value=2.53769e-29,
Score=319
Ontologies
GO
PANTHER
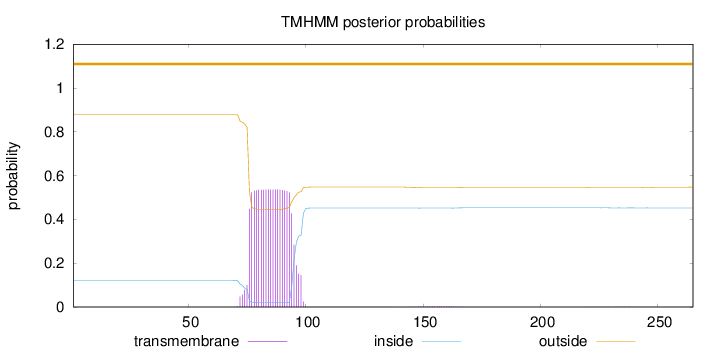
Topology
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.11768
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12189
outside
1 - 265
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
