Gene
KWMTBOMO16553 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004596
Annotation
PREDICTED:_Na(+)/H(+)_exchange_regulatory_cofactor_NHE-RF1_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.47
Sequence
CDS
ATGTCCGCGAACGGGTCGGCGGCCATCGAGCCACGACTCTGCCACGTGCGGAAGACGCCAGACTTCGACGGCTACGGATTTAATTTGCACGCAGAAAAAGGCAAACCCGGTCAGTACATCGGTAAGGTTGACGATGGCTCTCCGGCAGAGACCGCTGGCTTAAAACGCGGCGACCGCATCCTCGAAGTGAACGGCCACAGTATTGCCGGCGAGACCCACAAGCAAGTGGTGGCTCGCATCAAGGAGCGGCCCGATGACGCCGAGCTGCTGGTGATAGCACCGGCCCGGGGGACCAGATGCCCGATTTGGACGTGCCGGTGCCGGAAAAGACCCCTCCGCCTAGCGAAGCGCCCCGGGACAGTCCGTCGCCGACCGAACCTCCTAAGCTGA
Protein
MSANGSAAIEPRLCHVRKTPDFDGYGFNLHAEKGKPGQYIGKVDDGSPAETAGLKRGDRILEVNGHSIAGETHKQVVARIKERPDDAELLVIAPARGTRCPIWTCRCRKRPLRLAKRPGTVRRRPNLLS
Summary
Uniprot
H9J506
A0A3S2NPD1
A0A212F5U4
A0A2H1W8V1
A0A0N1IPZ4
A0A0N1IN07
+ More
A0A0L7L0T2 S4P182 N6TGH6 A0A084W4G7 A0A182YBR7 A0A2M4AUS0 A0A2M4AE36 A0A182FU83 A0A2M4BVP4 A0A2M4CT35 A0A2M4CRQ1 A0A2M4CRU3 A0A2M4CRN1 T1DFW4 A0A2M4BV76 A0A182ISA4 A0A182W272 A0A1Q3FH24 A0A1Q3FGQ8 B0XG18 A0A1Q3FH00 A0A1Q3FH40 A0A1Q3FGV6 A0A1Q3FGY7 A0A1Q3FHA8 A0A1Q3FGU3 A0A1Q3FGX6 A0A1Q3FH74 A0A182QAE0 A0A2M4AYF2 A0A2M4AC94 Q16K31 A0A2M4ACY0 A0A182XFR7 A0A1S4H843 A0A182HHT4 A0A182RY32 A0A182V3M0 Q7PV46 A0A1J1J6Q6 A0A1B6ITI6 A0A1B6H1L5 A0A182N9R2 E0W2S9 A0A1L8DKL0 V4B346 A0A1L8DKP0 R4WU03 A0A0N7ZBN2
A0A0L7L0T2 S4P182 N6TGH6 A0A084W4G7 A0A182YBR7 A0A2M4AUS0 A0A2M4AE36 A0A182FU83 A0A2M4BVP4 A0A2M4CT35 A0A2M4CRQ1 A0A2M4CRU3 A0A2M4CRN1 T1DFW4 A0A2M4BV76 A0A182ISA4 A0A182W272 A0A1Q3FH24 A0A1Q3FGQ8 B0XG18 A0A1Q3FH00 A0A1Q3FH40 A0A1Q3FGV6 A0A1Q3FGY7 A0A1Q3FHA8 A0A1Q3FGU3 A0A1Q3FGX6 A0A1Q3FH74 A0A182QAE0 A0A2M4AYF2 A0A2M4AC94 Q16K31 A0A2M4ACY0 A0A182XFR7 A0A1S4H843 A0A182HHT4 A0A182RY32 A0A182V3M0 Q7PV46 A0A1J1J6Q6 A0A1B6ITI6 A0A1B6H1L5 A0A182N9R2 E0W2S9 A0A1L8DKL0 V4B346 A0A1L8DKP0 R4WU03 A0A0N7ZBN2
Pubmed
EMBL
BABH01025768
RSAL01000017
RVE52892.1
AGBW02010136
OWR49098.1
ODYU01006767
+ More
SOQ48914.1 KQ460077 KPJ17854.1 KQ459444 KPJ00947.1 JTDY01003774 KOB69025.1 GAIX01009026 JAA83534.1 APGK01039170 KB740967 KB632399 ENN76883.1 ERL94629.1 ATLV01020332 KE525298 KFB45111.1 GGFK01011203 MBW44524.1 GGFK01005651 MBW38972.1 GGFJ01007860 MBW57001.1 GGFL01003820 MBW67998.1 GGFL01003819 MBW67997.1 GGFL01003817 MBW67995.1 GGFL01003818 MBW67996.1 GAMD01002996 JAA98594.1 GGFJ01007845 MBW56986.1 GFDL01008210 JAV26835.1 GFDL01008305 JAV26740.1 DS232981 EDS27176.1 GFDL01008262 JAV26783.1 GFDL01008196 JAV26849.1 GFDL01008235 JAV26810.1 GFDL01008224 JAV26821.1 GFDL01008121 JAV26924.1 GFDL01008296 JAV26749.1 GFDL01008238 JAV26807.1 GFDL01008139 JAV26906.1 AXCN02001804 GGFK01012454 MBW45775.1 GGFK01005080 MBW38401.1 CH477980 EAT34642.1 GGFK01005289 MBW38610.1 AAAB01008986 APCN01002441 EAA00462.5 CVRI01000074 CRL08101.1 GECU01017455 JAS90251.1 GECZ01026505 GECZ01018785 GECZ01008477 GECZ01003178 GECZ01003031 GECZ01001208 JAS43264.1 JAS50984.1 JAS61292.1 JAS66591.1 JAS66738.1 JAS68561.1 DS235879 EEB19935.1 GFDF01007071 JAV07013.1 KB199652 ESP04688.1 GFDF01007072 JAV07012.1 AK418192 BAN21407.1 GDRN01080866 JAI62120.1
SOQ48914.1 KQ460077 KPJ17854.1 KQ459444 KPJ00947.1 JTDY01003774 KOB69025.1 GAIX01009026 JAA83534.1 APGK01039170 KB740967 KB632399 ENN76883.1 ERL94629.1 ATLV01020332 KE525298 KFB45111.1 GGFK01011203 MBW44524.1 GGFK01005651 MBW38972.1 GGFJ01007860 MBW57001.1 GGFL01003820 MBW67998.1 GGFL01003819 MBW67997.1 GGFL01003817 MBW67995.1 GGFL01003818 MBW67996.1 GAMD01002996 JAA98594.1 GGFJ01007845 MBW56986.1 GFDL01008210 JAV26835.1 GFDL01008305 JAV26740.1 DS232981 EDS27176.1 GFDL01008262 JAV26783.1 GFDL01008196 JAV26849.1 GFDL01008235 JAV26810.1 GFDL01008224 JAV26821.1 GFDL01008121 JAV26924.1 GFDL01008296 JAV26749.1 GFDL01008238 JAV26807.1 GFDL01008139 JAV26906.1 AXCN02001804 GGFK01012454 MBW45775.1 GGFK01005080 MBW38401.1 CH477980 EAT34642.1 GGFK01005289 MBW38610.1 AAAB01008986 APCN01002441 EAA00462.5 CVRI01000074 CRL08101.1 GECU01017455 JAS90251.1 GECZ01026505 GECZ01018785 GECZ01008477 GECZ01003178 GECZ01003031 GECZ01001208 JAS43264.1 JAS50984.1 JAS61292.1 JAS66591.1 JAS66738.1 JAS68561.1 DS235879 EEB19935.1 GFDF01007071 JAV07013.1 KB199652 ESP04688.1 GFDF01007072 JAV07012.1 AK418192 BAN21407.1 GDRN01080866 JAI62120.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000019118 UP000030742 UP000030765 UP000076408 UP000069272 UP000075880 UP000075920 UP000002320 UP000075886 UP000008820 UP000076407 UP000075840 UP000075900 UP000075903 UP000007062 UP000183832 UP000075884 UP000009046 UP000030746
UP000019118 UP000030742 UP000030765 UP000076408 UP000069272 UP000075880 UP000075920 UP000002320 UP000075886 UP000008820 UP000076407 UP000075840 UP000075900 UP000075903 UP000007062 UP000183832 UP000075884 UP000009046 UP000030746
PRIDE
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
H9J506
A0A3S2NPD1
A0A212F5U4
A0A2H1W8V1
A0A0N1IPZ4
A0A0N1IN07
+ More
A0A0L7L0T2 S4P182 N6TGH6 A0A084W4G7 A0A182YBR7 A0A2M4AUS0 A0A2M4AE36 A0A182FU83 A0A2M4BVP4 A0A2M4CT35 A0A2M4CRQ1 A0A2M4CRU3 A0A2M4CRN1 T1DFW4 A0A2M4BV76 A0A182ISA4 A0A182W272 A0A1Q3FH24 A0A1Q3FGQ8 B0XG18 A0A1Q3FH00 A0A1Q3FH40 A0A1Q3FGV6 A0A1Q3FGY7 A0A1Q3FHA8 A0A1Q3FGU3 A0A1Q3FGX6 A0A1Q3FH74 A0A182QAE0 A0A2M4AYF2 A0A2M4AC94 Q16K31 A0A2M4ACY0 A0A182XFR7 A0A1S4H843 A0A182HHT4 A0A182RY32 A0A182V3M0 Q7PV46 A0A1J1J6Q6 A0A1B6ITI6 A0A1B6H1L5 A0A182N9R2 E0W2S9 A0A1L8DKL0 V4B346 A0A1L8DKP0 R4WU03 A0A0N7ZBN2
A0A0L7L0T2 S4P182 N6TGH6 A0A084W4G7 A0A182YBR7 A0A2M4AUS0 A0A2M4AE36 A0A182FU83 A0A2M4BVP4 A0A2M4CT35 A0A2M4CRQ1 A0A2M4CRU3 A0A2M4CRN1 T1DFW4 A0A2M4BV76 A0A182ISA4 A0A182W272 A0A1Q3FH24 A0A1Q3FGQ8 B0XG18 A0A1Q3FH00 A0A1Q3FH40 A0A1Q3FGV6 A0A1Q3FGY7 A0A1Q3FHA8 A0A1Q3FGU3 A0A1Q3FGX6 A0A1Q3FH74 A0A182QAE0 A0A2M4AYF2 A0A2M4AC94 Q16K31 A0A2M4ACY0 A0A182XFR7 A0A1S4H843 A0A182HHT4 A0A182RY32 A0A182V3M0 Q7PV46 A0A1J1J6Q6 A0A1B6ITI6 A0A1B6H1L5 A0A182N9R2 E0W2S9 A0A1L8DKL0 V4B346 A0A1L8DKP0 R4WU03 A0A0N7ZBN2
PDB
2HE4
E-value=1.37333e-20,
Score=239
Ontologies
GO
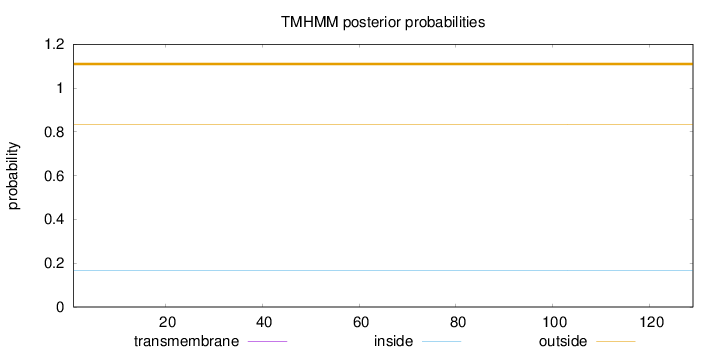
Topology
Length:
129
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00015
Exp number, first 60 AAs:
0
Total prob of N-in:
0.16653
outside
1 - 129
Population Genetic Test Statistics
Pi
24.81665
Theta
33.562787
Tajima's D
-0.761407
CLR
0
CSRT
0.184490775461227
Interpretation
Uncertain
