Gene
KWMTBOMO16543
Pre Gene Modal
BGIBMGA013407
Annotation
PREDICTED:_dual_serine/threonine_and_tyrosine_protein_kinase-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.763
Sequence
CDS
ATGGCCGGTGGGTGTGCGTGGTCGCGACGTGTGGGCGCCCGAACGAGGACTCTTAGAGGATTAGTTCGGGACACACAGCGGGCTCTGAACGACGTCTGCGATATATTGAATTATGATCCGGATATCCTGGTAACAGTTCAAGATATAGGCAACCAGTTAGCGAGAAGCACAGCTATCGTAGTACTAGGGAGCACGCCGTCGGCCAAGGCTAGACTTTTGCACTGCTTGTTGGGACGACAGCTGCTGCCGGATTCTCCGCCGAGAGGATGCAGATGGCTTCGTATCCAGTACGGGAGCACAACCCAGGTCCACCTGACCCTCGGCAACTCAGAGTTCGAGCTCGTCGAAGAGTTGGAGTGCAACAAGCGTCCCTGGGAGACGCTGCCCGTTGAGGATCTCCTACGACGAGACAACGCCGACGCCTCAACGATACTAGAAGTGGAAATCAACAATCAATTCCTAAGAGACGGCCTCAAGATCATCATACCTCCCGACATCAGCGTCGAGCCAGGCCTAGCAGACCACCTGACCCTGAAGAAGCTCCACCATGAACTGTACACGAAAAGAGAAAGCATCTTGAGGAGCATCAACCCGGTATACCTGTACGGTATCGACAGAATGGGAAAGAACATCTTCAGCGATAATATAATGAATTCGCTTGTGGTGTCGAAGAGCGAAGAAGACTTTTGGGACAGTTTCAATCTGTACTGCATGAGCAAGAGTGAGAGGGAGAGAGAGAAGACTGAGATCGTTATAGATAGAGAGGACAGAGAGTGGAATTCAACGAGGGAGCCTTGTGTTTTCAGTGGGGAGAACTGCCTGGACCTGCACCAAATCAAGGAGATAAATCCGAACGCTCAAGTGATGTTCGTATTGTTCTCGGACAGCGCGGAGACGTGCGGCGCGGGGCGGGGGGGCGGCGCCGCGCGGCTGCTGCTCGACGTGGAGGCGACGGAGTCCACCGAGGGCTCCGTGTGCTGCGACGAGCTGCCGGAGAGCGACCAGGGCCTGGTCCAGATACGGAATAATAACGGGTCTCCGACATCGAAAGCTCAGGACGACAGACGACTGCACGAAGCACAAATAGCTTTTTTGAACGAGCTTCTCGACCAGTGGGAGCTTATGTCGTCCCGGCCGCAAAAGCACCAAGTCAAGAGCCAGTGGATGATCCTGAACGAGCTGGCGGTGCACCGCGGGGGCGAGGGCGGGGCGGGCGGCGCGCGGGGGGCGGGGTCGCGCACGGCGCTGCTGGGGGCCGTCGTGCGCTTCGCCACTGACACCGCGCTGGCCGACCTGCTGCAGCGCGCCACCAGGCTCGGCGAGATCCACGTGAAGCTGCTCCAGCAGTTCATCCTGGCCTCGTACGACATGGCCCGAGAGCTGCAGGTGGTGCCCAAGAAGATCCAGTACGTGGCACGGCAGGAGCAGCAGCTGTATGAGACTATACACGAGAAGTTCACCGAGGGTGACAAGAAGCACGAGCTGCTGCGGATAATGCAGGACGTGCTGCAAGAGATGAAGCACGAGATCAACAACATGGACTGGAGCGTCGACGACCTGCCGTGCCACCAGGACCACCGCTTCACCGTGTCCAGCTCTACGTTCTACTCGGCGCGGCCGAGTCCGGAGCACCCGCGCGACTGGTCGCGGAGCCCTGACCCGTCGGACGGCGAGGAGGCCGTCTCCTACGACACCTACAACTTCGACGACTACGAGATCATCCAGCAGGGCCTGGATACCTTCGGGGACTCACTTCCGATGAGGGTCCGACCCGAGGAGGACGCGTCCAAGTTCAACAACATCAAGATCGAGAATGAATACGAGTCGTGCTCCGTGTCGTCGGCGCGCAGCGGCTGGGCGGTGGCGACCGGCAGTGACGTCACCACCTGCACCAACAACACCAACGTCTCCATCAAGCAGGCGTCGCTGGACGTGCAGCAGACCGTGCTATCGAAGCTGAGCCGAAAAATTAGTTTGAAACTGGTGAAGTTCGTGGATTGCCTCAAGAACACGTACTTCGGTACGCTGCAAAGATGTCTAGAATCCCTAGAATCATCTTGCAGACAGGAGTTGGGCGGTCTACCGGCCAGTGAAGCAATGAGGCAGCTGCTCTCGGTTGCCTGGCAGGTGGATCTCCAGCCCTGTGCCTCCTTCTCTGTACTGAGGACTCTAATCGAATCACTGAGACGTCTTTTGCATAAATTGAAGATAGTGAGCGGTGAAGATGATGACGCGTGTTGCGTGCTGAGTCCGGTGTGGAGGCGCCACGTGGCGATGCACGCCGCGCACTCGCTCAACCCGCACCGCCTCGCCAGGCTCATATCCACGCAGATCCTAGAGAAGCTAAGCACGGCTCACGAACGCTATCAATCGGCGCTGGCGTCCCTCGAGGCGGCCCTGACGGGGCGCCTCCACCACACCGAGGACGTGAAGCTGGCCATCAGGAAGAAACACGCCCCGTCCTTCGCCAGACTCTGCCTCGAGAGCACGTCCCTGTGCGACATGCTGATGCACGGCGTTCCGGAGCTGGGCAGGGAGATCGGTCGCGGGCAGTACGGCGTGGTTTACGCCGTCCGCGGCAGCTGGGGGGGCCACTCGCCGGTCGCGGTCAAGTCGGTGCTGCCCACGGACGAGCGCCATTACCACGAGCTGGCCATGGAATTCTTTTATACTAGGAGTATACCGGCGCACCCCCGCATAGTCCGCCTGTACGGGTCGATAGTACAGCGCCAGAGTTCTGGGCCCAGCGTGCTGCTGGTGTCCGAGAGGAAATGTCGGGATCTGCACGCGGCCGTCAGGGCGAAACTCCCCTTCTCCACTCGGATGAGGATCAGCTGCGATATCGTCGACGGCATCCGCTACCTGCACTCGCTGGGTCTGGTGCACCGCGACATCAAGACGAAGAACGTCCTCCTGGACGGGGCGGACCGCGCGGCGCTGTCGGACCTCGGCTTCTGCACGGCCGGCGCGCTCATGTCGGGCTCCGTGGTGGGCACGCCGGTGCACATGGCGCCCGAGCTGCTGGCCGGGGACTACCACGCCTCCGTCGACGTGTACGCTTTCGGGATCTTGTTCTGGTACGTGTGCGCTGGTGACATAAAGCTACCGTCAGCTTTCGAAACTTTCCAAAACAAGGAACAGCTTTGGAGTAAGGTTAAAAGAGGTCTGAGACCTGAACGTCTACCTCAGTTCTCGGACGAGTGCTGGGAGATCATGGAGTCGTGCTGGGCCTCCGAGCCCTCCCAGCGGGCGCTCCTCGGGGACGTGCAGAACAAGCTGGAGAGGATCTTGGAGCAGGCTCTAAAGGACGAAAAGTCTCCTGGCTACCTCGCCCCGGACGAGCCAGACCTTAACGACGACTAG
Protein
MAGGCAWSRRVGARTRTLRGLVRDTQRALNDVCDILNYDPDILVTVQDIGNQLARSTAIVVLGSTPSAKARLLHCLLGRQLLPDSPPRGCRWLRIQYGSTTQVHLTLGNSEFELVEELECNKRPWETLPVEDLLRRDNADASTILEVEINNQFLRDGLKIIIPPDISVEPGLADHLTLKKLHHELYTKRESILRSINPVYLYGIDRMGKNIFSDNIMNSLVVSKSEEDFWDSFNLYCMSKSEREREKTEIVIDREDREWNSTREPCVFSGENCLDLHQIKEINPNAQVMFVLFSDSAETCGAGRGGGAARLLLDVEATESTEGSVCCDELPESDQGLVQIRNNNGSPTSKAQDDRRLHEAQIAFLNELLDQWELMSSRPQKHQVKSQWMILNELAVHRGGEGGAGGARGAGSRTALLGAVVRFATDTALADLLQRATRLGEIHVKLLQQFILASYDMARELQVVPKKIQYVARQEQQLYETIHEKFTEGDKKHELLRIMQDVLQEMKHEINNMDWSVDDLPCHQDHRFTVSSSTFYSARPSPEHPRDWSRSPDPSDGEEAVSYDTYNFDDYEIIQQGLDTFGDSLPMRVRPEEDASKFNNIKIENEYESCSVSSARSGWAVATGSDVTTCTNNTNVSIKQASLDVQQTVLSKLSRKISLKLVKFVDCLKNTYFGTLQRCLESLESSCRQELGGLPASEAMRQLLSVAWQVDLQPCASFSVLRTLIESLRRLLHKLKIVSGEDDDACCVLSPVWRRHVAMHAAHSLNPHRLARLISTQILEKLSTAHERYQSALASLEAALTGRLHHTEDVKLAIRKKHAPSFARLCLESTSLCDMLMHGVPELGREIGRGQYGVVYAVRGSWGGHSPVAVKSVLPTDERHYHELAMEFFYTRSIPAHPRIVRLYGSIVQRQSSGPSVLLVSERKCRDLHAAVRAKLPFSTRMRISCDIVDGIRYLHSLGLVHRDIKTKNVLLDGADRAALSDLGFCTAGALMSGSVVGTPVHMAPELLAGDYHASVDVYAFGILFWYVCAGDIKLPSAFETFQNKEQLWSKVKRGLRPERLPQFSDECWEIMESCWASEPSQRALLGDVQNKLERILEQALKDEKSPGYLAPDEPDLNDD
Summary
EMBL
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
5AMN
E-value=1.84274e-18,
Score=232
Ontologies
GO
Topology
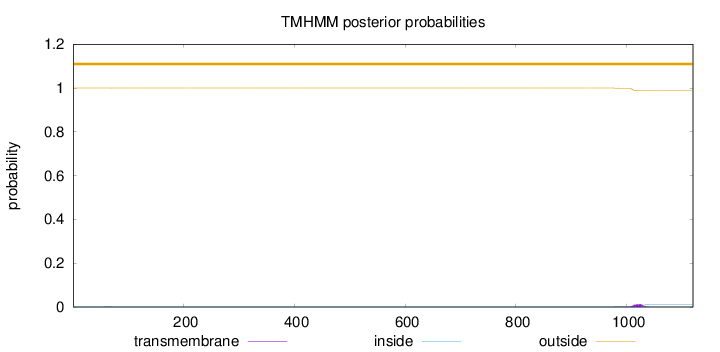
Length:
1120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2579
Exp number, first 60 AAs:
0.00097
Total prob of N-in:
0.00017
outside
1 - 1120
Population Genetic Test Statistics
Pi
25.193447
Theta
29.349663
Tajima's D
-0.780946
CLR
0.001618
CSRT
0.178691065446728
Interpretation
Uncertain
