Gene
KWMTBOMO16539
Pre Gene Modal
BGIBMGA005643
Annotation
PREDICTED:_WAS_protein_family_homolog_1-like_[Amyelois_transitella]
Full name
WASH complex subunit 1
+ More
WAS protein family homolog 2
WAS protein family homolog 2
Alternative Name
WAS protein family homolog 1
CXYorf1-like protein on chromosome 2
Protein FAM39B
CXYorf1-like protein on chromosome 9
Protein FAM39E
CXYorf1-like protein on chromosome 2
Protein FAM39B
CXYorf1-like protein on chromosome 9
Protein FAM39E
Location in the cell
Cytoplasmic Reliability : 1.785 Nuclear Reliability : 1.966
Sequence
CDS
ATGGAAGGTATTTACAAAATTAATCTTGTGCCAAGCGACTTGAGCACTGAAGAAGCCATCCTTCAGATTGCTGACACTATTGATAATCTCAATGGGATAGTGGAAGACGTGTTCTCTCGCTTAACGCGGCGAATCAAGCAGAATGTAGATAAAACCGCAAAATTGAATGAACGCATCGAAAACTCTAGGCTTAAAGTTGAAAAACTGACTGGAATGCAGAGAGCCATCAAAGTGTTCTCGAGTGCAAAGTACCCGTCGTCAATAACGCACGAGCATTATCGATCCGTCTTTGATTTGGAAGGCTATCAACATGTGCCTAAAAATGTTAAACTGAGTGCTAAAACTCGGGGGCCATCTGATGATAAAAGAATACAGGAGAAGCTACACTTCTATCATGTTAAAGTTGCGGAACCTAATTCACTCAAGCCCGATGAAGATTTTAGTTTGAACGCTGTTATTGATAACCTGGAAACCATCGGTGAACTTTTAGTATTTAACACCGATGAAAGCCCTTACTTCGGTGTTACCTCTAAAACACCAACCTACAAACCGGTCATCAAGAAATCGAATATTGAAAATAAGAGTCTAATAGAAGCGGCACCTTCGTCTATAATGAAGAAAGACGTCTTGAAGCGCGAAGTAGACGAATACATGTACGCACCTGGGATGGGAATGGTACCCGAAATGGAGATGCCGTTGGATTTACCGCATCTTCCAGGAATCGCTGAGGACATACAATATACTATCGCGACTGAAGGGTCTATAGCCCCGTCTGTCGTGACGTCACCGGTCACGCCGATCATCGCCATTCCTGAGGTGGAATTACCAGAGTTGCCCGATCTAAAGGAACTACCTGACGTGAAACCAGAGCCCGTGGCTATACCAGATATACCGGACGCTCCTCCGTCACCGGTTCCTGTTAGTGTGCAAGCGTTGCCACCACCGCCGCCCCCGCCACCGCCTCCGCCCCCGCCGCCGCCTATGAAGGAAGTAGAGCTCACACCATCAAAACGGTAA
Protein
MEGIYKINLVPSDLSTEEAILQIADTIDNLNGIVEDVFSRLTRRIKQNVDKTAKLNERIENSRLKVEKLTGMQRAIKVFSSAKYPSSITHEHYRSVFDLEGYQHVPKNVKLSAKTRGPSDDKRIQEKLHFYHVKVAEPNSLKPDEDFSLNAVIDNLETIGELLVFNTDESPYFGVTSKTPTYKPVIKKSNIENKSLIEAAPSSIMKKDVLKREVDEYMYAPGMGMVPEMEMPLDLPHLPGIAEDIQYTIATEGSIAPSVVTSPVTPIIAIPEVELPELPDLKELPDVKPEPVAIPDIPDAPPSPVPVSVQALPPPPPPPPPPPPPPPMKEVELTPSKR
Summary
Description
Acts as a nucleation-promoting factor at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting. Its assembly in the WASH core complex seems to inhibit its NPF activity and is required for its membrane targeting.
Acts as a nucleation-promoting factor (NPF) at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting. Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting. Involved in endocytic trafficking of EGF. Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration. In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T-cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation. Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Acts as a nucleation-promoting factor at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting. Involved in endocytic trafficking of EGF. Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting. Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration. In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T-cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation. Involved in Arp2/3 complex-dependent actin assembly driving Salmonella typhimurium invasion independent of ruffling. Involved in the exocytosis of MMP14 leading to matrix remodeling during invasive migration and implicating late endosome-to-plasma membrane tubular connections and cooperation with the exocyst complex. Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Acts as a nucleation-promoting factor (NPF) at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting (PubMed:19922874, PubMed:19922875, PubMed:20498093, PubMed:23452853). Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting (PubMed:20498093). Involved in endocytic trafficking of EGF (By similarity). Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration (PubMed:22114305). In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation (By similarity). Involved in Arp2/3 complex-dependent actin assembly driving Salmonella typhimurium invasion independent of ruffling. Involved in the exocytosis of MMP14 leading to matrix remodeling during invasive migration and implicating late endosome-to-plasma membrane tubular connections and cooperation with the exocyst complex (PubMed:24344185). Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Acts as a nucleation-promoting factor (NPF) at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting. Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting. Involved in endocytic trafficking of EGF. Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration. In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T-cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation. Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Acts as a nucleation-promoting factor at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting. Involved in endocytic trafficking of EGF. Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting. Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration. In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T-cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation. Involved in Arp2/3 complex-dependent actin assembly driving Salmonella typhimurium invasion independent of ruffling. Involved in the exocytosis of MMP14 leading to matrix remodeling during invasive migration and implicating late endosome-to-plasma membrane tubular connections and cooperation with the exocyst complex. Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Acts as a nucleation-promoting factor (NPF) at the surface of endosomes, where it recruits and activates the Arp2/3 complex to induce actin polymerization, playing a key role in the fission of tubules that serve as transport intermediates during endosome sorting (PubMed:19922874, PubMed:19922875, PubMed:20498093, PubMed:23452853). Its assembly in the WASH core complex seems to inhibit its NPF activity and via WASHC2 is required for its membrane targeting (PubMed:20498093). Involved in endocytic trafficking of EGF (By similarity). Involved in transferrin receptor recycling. Regulates the trafficking of endosomal alpha5beta1 integrin to the plasma membrane and involved in invasive cell migration (PubMed:22114305). In T-cells involved in endosome-to-membrane recycling of receptors including T-cell receptor (TCR), CD28 and ITGAL; proposed to be implicated in T cell proliferation and effector function. In dendritic cells involved in endosome-to-membrane recycling of major histocompatibility complex (MHC) class II probably involving retromer and subsequently allowing antigen sampling, loading and presentation during T-cell activation (By similarity). Involved in Arp2/3 complex-dependent actin assembly driving Salmonella typhimurium invasion independent of ruffling. Involved in the exocytosis of MMP14 leading to matrix remodeling during invasive migration and implicating late endosome-to-plasma membrane tubular connections and cooperation with the exocyst complex (PubMed:24344185). Involved in negative regulation of autophagy independently from its role in endosomal sorting by inhibiting BECN1 ubiquitination to inactivate PIK3C3/Vps34 activity (By similarity).
Subunit
Component of the WASH complex.
Component of the WASH core complex also described as WASH regulatory complex SHRC composed of WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. The WASH core complex associates with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB); the assembly has been initially described as WASH complex (PubMed:20498093). Interacts (via WHD1 region) with WASHC2; the interaction is direct. Interacts with alpha-tubulin. Interacts with BECN1; WASHC1 and AMBRA1 can competetively interact with BECN1. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2) (By similarity). Interacts with TBC1D23 (By similarity).
Component of the WASH core complex also described as WASH regulatory complex (SHRC) composed of WASH (WASHC1, WASH2P or WASH3P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5. The WASH core complex associates with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB) in a transient or substoichiometric manner which was initially described as WASH complex. Interacts (via WHD1 region) with WASHC2C; the interaction is direct. Interacts with alpha-tubulin. Interacts with BECN1; WASHC1 and AMBRA1 can competetively interact with BECN1. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2). Interacts with EXOC1, EXOC4, EXOC8; in MMP14-positive endosomes in breast tumor cells; indicative for an association with the exocyst complex (By similarity).
Component of the WASH core complex also described as WASH regulatory complex (SHRC) composed of WASH (WASHC1, WASH2P or WASH3P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5. The WASH core complex associates via WASHC2 with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB) in a transient or substoichiometric manner which was initially described as WASH complex (PubMed:19922875, PubMed:20498093). Interacts (via WHD1 region) with WASHC2C; the interaction is direct (PubMed:19922874). Interacts with VPS35; mediates the association with the retromer CSC complex. Interacts with FKBP15. Interacts with alpha-tubulin. Interacts with BECN1; this interaction can be competed out by AMBRA1 binding. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2) (By similarity). Interacts with EXOC1, EXOC4, EXOC8; in MMP14-positive endosomes in breast tumor cells; indicative for an association with the exocyst complex (PubMed:24344185). Interacts with TBC1D23 (PubMed:29084197).
Component of the WASH core complex also described as WASH regulatory complex SHRC composed of WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. The WASH core complex associates with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB); the assembly has been initially described as WASH complex (PubMed:20498093). Interacts (via WHD1 region) with WASHC2; the interaction is direct. Interacts with alpha-tubulin. Interacts with BECN1; WASHC1 and AMBRA1 can competetively interact with BECN1. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2) (By similarity). Interacts with TBC1D23 (By similarity).
Component of the WASH core complex also described as WASH regulatory complex (SHRC) composed of WASH (WASHC1, WASH2P or WASH3P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5. The WASH core complex associates with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB) in a transient or substoichiometric manner which was initially described as WASH complex. Interacts (via WHD1 region) with WASHC2C; the interaction is direct. Interacts with alpha-tubulin. Interacts with BECN1; WASHC1 and AMBRA1 can competetively interact with BECN1. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2). Interacts with EXOC1, EXOC4, EXOC8; in MMP14-positive endosomes in breast tumor cells; indicative for an association with the exocyst complex (By similarity).
Component of the WASH core complex also described as WASH regulatory complex (SHRC) composed of WASH (WASHC1, WASH2P or WASH3P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5. The WASH core complex associates via WASHC2 with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB) in a transient or substoichiometric manner which was initially described as WASH complex (PubMed:19922875, PubMed:20498093). Interacts (via WHD1 region) with WASHC2C; the interaction is direct (PubMed:19922874). Interacts with VPS35; mediates the association with the retromer CSC complex. Interacts with FKBP15. Interacts with alpha-tubulin. Interacts with BECN1; this interaction can be competed out by AMBRA1 binding. Interacts with BLOC1S2; may associate with the BLOC-1 complex. Interacts with tubulin gamma chain (TUBG1 or TUBG2) (By similarity). Interacts with EXOC1, EXOC4, EXOC8; in MMP14-positive endosomes in breast tumor cells; indicative for an association with the exocyst complex (PubMed:24344185). Interacts with TBC1D23 (PubMed:29084197).
Miscellaneous
WASH genes duplicated to multiple chromosomal ends during primate evolution, with highest copy number reached in humans, whose WASH repertoires probably vary extensively among individuals (PubMed:18159949). It is therefore difficult to determine which gene is functional or not. The telomeric region of chromosome 9p is paralogous to the pericentromeric regions of chromosome 9 as well as to 2q. Paralogous regions contain 7 transcriptional units. Duplicated WASH genes are also present in the Xq/Yq pseudoautosomal region, as well as on chromosome 1 and 15. The chromosome 16 copy seems to be a pseudogene.
Similarity
Belongs to the WASH1 family.
Keywords
Actin-binding
Complete proteome
Endosome
Membrane
Protein transport
Reference proteome
Transport
Cytoplasm
Cytoplasmic vesicle
Cytoskeleton
Isopeptide bond
Ubl conjugation
Feature
chain WASH complex subunit 1
Uniprot
H9J801
A0A1E1W144
A0A2A4JDC2
A0A2H1W478
D0AB84
A0A194QS37
+ More
L7X0Q8 A0A194QGQ8 A0A212EP92 A0A0L7LB49 A0A067QUW1 A0A2J7QEZ0 A0A2J7QEY0 A0A0L8FGQ0 A0A1Y1K9I7 A0A1W4WAS5 W4XZ20 C3XT97 Q28DN4 K1QHN6 A7S4U2 T1J4E9 Q5U4A3 A0A1W2W4Z8 G7PN26 A0A2K6L8U9 A0A2K6P244 A0A2K6BTR6 G7NAZ7 A0A2K5KYP6 A0A096MNA7 A0A2K5ZHL7 H2P7E0 A0A0D9RIB8 G1QMF0 A0A2G8L2J7 A0A2K5J8N9 U4UPZ8 F7ACG4 A0A286XH06 K7BSJ4 G1U687 A7Z063 U4UT95 A0A2C9JEA9 N6U7W4 G3RVA7 I3MT98 G5B7J2 A0A3Q2H2I4 F7E3G3 A0A3P4PLA7 A0A0P7YEC5 F6QVX5 A0A1W5AZA3 A0A0P6JAG2 A0A3B3ST62 L9LCK4 G9KXK8 A0A2Y9KYL4 G3WP97 A0A2U3WS11 M3Z2V3 A0A2P6LBT2 A0A341AI29 A0A3Q7NWB8 M3X1I8 A0A3Q7ML03 A0A2Y9NJC0 A0A2Y9NU53 A0A341AGR2 A0A341AGQ5 A0A2Y9NNU5 Q6VEQ5 R7V9U1 F1SK83 A0A341AFV8 A0A287BQQ1 A0A2Y9NDJ0 A0A341ADS1 A0A2Y9NG41 A0A2Y9H4C6 A0A2U3YE77 A8K0Z3 A0A2U3YDX0 A0A3Q7T8B8 A0A1B0GJ14 E2QXP1 A0A3Q7RWZ3 G1LDI4 G1LDG8 A0A3Q7RZW7 A0A383Z564 A0A2G9RWY6 A0A3Q7WAA2 A0A384D8G4 A0A060WYS1 A0A383Z5G9 A0A383Z5D1
L7X0Q8 A0A194QGQ8 A0A212EP92 A0A0L7LB49 A0A067QUW1 A0A2J7QEZ0 A0A2J7QEY0 A0A0L8FGQ0 A0A1Y1K9I7 A0A1W4WAS5 W4XZ20 C3XT97 Q28DN4 K1QHN6 A7S4U2 T1J4E9 Q5U4A3 A0A1W2W4Z8 G7PN26 A0A2K6L8U9 A0A2K6P244 A0A2K6BTR6 G7NAZ7 A0A2K5KYP6 A0A096MNA7 A0A2K5ZHL7 H2P7E0 A0A0D9RIB8 G1QMF0 A0A2G8L2J7 A0A2K5J8N9 U4UPZ8 F7ACG4 A0A286XH06 K7BSJ4 G1U687 A7Z063 U4UT95 A0A2C9JEA9 N6U7W4 G3RVA7 I3MT98 G5B7J2 A0A3Q2H2I4 F7E3G3 A0A3P4PLA7 A0A0P7YEC5 F6QVX5 A0A1W5AZA3 A0A0P6JAG2 A0A3B3ST62 L9LCK4 G9KXK8 A0A2Y9KYL4 G3WP97 A0A2U3WS11 M3Z2V3 A0A2P6LBT2 A0A341AI29 A0A3Q7NWB8 M3X1I8 A0A3Q7ML03 A0A2Y9NJC0 A0A2Y9NU53 A0A341AGR2 A0A341AGQ5 A0A2Y9NNU5 Q6VEQ5 R7V9U1 F1SK83 A0A341AFV8 A0A287BQQ1 A0A2Y9NDJ0 A0A341ADS1 A0A2Y9NG41 A0A2Y9H4C6 A0A2U3YE77 A8K0Z3 A0A2U3YDX0 A0A3Q7T8B8 A0A1B0GJ14 E2QXP1 A0A3Q7RWZ3 G1LDI4 G1LDG8 A0A3Q7RZW7 A0A383Z564 A0A2G9RWY6 A0A3Q7WAA2 A0A384D8G4 A0A060WYS1 A0A383Z5G9 A0A383Z5D1
Pubmed
19121390
26354079
23674305
22118469
26227816
24845553
+ More
28004739 18563158 22992520 17615350 22002653 25362486 25319552 29023486 23537049 12481130 15114417 21993624 20498093 15562597 21993625 19892987 17495919 29240929 23385571 23236062 21709235 17975172 15815621 15233989 10655549 18159949 23254933 14702039 15164053 19922874 19922875 22114305 23452853 24344185 29084197 16341006 20010809 24755649
28004739 18563158 22992520 17615350 22002653 25362486 25319552 29023486 23537049 12481130 15114417 21993624 20498093 15562597 21993625 19892987 17495919 29240929 23385571 23236062 21709235 17975172 15815621 15233989 10655549 18159949 23254933 14702039 15164053 19922874 19922875 22114305 23452853 24344185 29084197 16341006 20010809 24755649
EMBL
BABH01020206
GDQN01010433
JAT80621.1
NWSH01001832
PCG69961.1
ODYU01006205
+ More
SOQ47848.1 FP102340 CBH09279.1 KQ461155 KPJ08302.1 KC469893 AGC92689.2 KQ459232 KPJ02631.1 AGBW02013517 OWR43294.1 JTDY01001858 KOB72702.1 KK852908 KDR13973.1 NEVH01015305 PNF27145.1 PNF27144.1 KQ432990 KOF62581.1 GEZM01093099 JAV56345.1 AAGJ04120810 GG666462 EEN68686.1 CR855419 BC123023 JH816276 EKC30659.1 DS469580 EDO41211.1 JH431845 BC085201 CM001288 EHH55871.1 JU332380 JV045823 CM001265 AFE76135.1 AFI35894.1 EHH22444.1 AHZZ02001504 ABGA01235378 AQIB01143913 AQIB01143914 ADFV01077923 ADFV01077924 MRZV01000247 PIK54489.1 KI210047 ERL96144.1 EAAA01002669 AAKN02038674 GABF01005071 JAA17074.1 AAGW02051487 BC153261 KI207928 ERL95758.1 APGK01045630 APGK01045631 KB741037 ENN74672.1 AGTP01074412 JH168787 EHB05253.1 CYRY02034003 VCX10606.1 JARO02006858 KPP64679.1 GEBF01004127 JAN99505.1 KB320463 ELW71462.1 JP021039 AES09637.1 AEFK01193797 AEFK01193798 AEFK01193799 AEYP01081943 MWRG01000475 PRD36026.1 AANG04001873 AL078621 AY341936 AMQN01005391 KB295903 ELU12510.1 AEMK02000037 AK289708 AL928970 AJWK01019202 AAEX03015316 ACTA01137259 KV930729 PIO31721.1 FR904704 CDQ70209.1
SOQ47848.1 FP102340 CBH09279.1 KQ461155 KPJ08302.1 KC469893 AGC92689.2 KQ459232 KPJ02631.1 AGBW02013517 OWR43294.1 JTDY01001858 KOB72702.1 KK852908 KDR13973.1 NEVH01015305 PNF27145.1 PNF27144.1 KQ432990 KOF62581.1 GEZM01093099 JAV56345.1 AAGJ04120810 GG666462 EEN68686.1 CR855419 BC123023 JH816276 EKC30659.1 DS469580 EDO41211.1 JH431845 BC085201 CM001288 EHH55871.1 JU332380 JV045823 CM001265 AFE76135.1 AFI35894.1 EHH22444.1 AHZZ02001504 ABGA01235378 AQIB01143913 AQIB01143914 ADFV01077923 ADFV01077924 MRZV01000247 PIK54489.1 KI210047 ERL96144.1 EAAA01002669 AAKN02038674 GABF01005071 JAA17074.1 AAGW02051487 BC153261 KI207928 ERL95758.1 APGK01045630 APGK01045631 KB741037 ENN74672.1 AGTP01074412 JH168787 EHB05253.1 CYRY02034003 VCX10606.1 JARO02006858 KPP64679.1 GEBF01004127 JAN99505.1 KB320463 ELW71462.1 JP021039 AES09637.1 AEFK01193797 AEFK01193798 AEFK01193799 AEYP01081943 MWRG01000475 PRD36026.1 AANG04001873 AL078621 AY341936 AMQN01005391 KB295903 ELU12510.1 AEMK02000037 AK289708 AL928970 AJWK01019202 AAEX03015316 ACTA01137259 KV930729 PIO31721.1 FR904704 CDQ70209.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000027135 UP000235965 UP000053454 UP000192223 UP000007110 UP000001554 UP000008143 UP000005408 UP000001593 UP000009130 UP000233180 UP000233200 UP000233120 UP000233060 UP000028761 UP000233140 UP000001595 UP000029965 UP000001073 UP000230750 UP000233080 UP000030742 UP000008144 UP000005447 UP000001811 UP000009136 UP000076420 UP000019118 UP000001519 UP000005215 UP000006813 UP000002281 UP000034805 UP000192224 UP000002280 UP000261540 UP000011518 UP000248482 UP000007648 UP000245340 UP000000715 UP000252040 UP000286641 UP000011712 UP000248483 UP000005640 UP000014760 UP000008227 UP000248481 UP000245341 UP000286640 UP000092461 UP000002254 UP000008912 UP000261681 UP000286642 UP000261680 UP000193380
UP000027135 UP000235965 UP000053454 UP000192223 UP000007110 UP000001554 UP000008143 UP000005408 UP000001593 UP000009130 UP000233180 UP000233200 UP000233120 UP000233060 UP000028761 UP000233140 UP000001595 UP000029965 UP000001073 UP000230750 UP000233080 UP000030742 UP000008144 UP000005447 UP000001811 UP000009136 UP000076420 UP000019118 UP000001519 UP000005215 UP000006813 UP000002281 UP000034805 UP000192224 UP000002280 UP000261540 UP000011518 UP000248482 UP000007648 UP000245340 UP000000715 UP000252040 UP000286641 UP000011712 UP000248483 UP000005640 UP000014760 UP000008227 UP000248481 UP000245341 UP000286640 UP000092461 UP000002254 UP000008912 UP000261681 UP000286642 UP000261680 UP000193380
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J801
A0A1E1W144
A0A2A4JDC2
A0A2H1W478
D0AB84
A0A194QS37
+ More
L7X0Q8 A0A194QGQ8 A0A212EP92 A0A0L7LB49 A0A067QUW1 A0A2J7QEZ0 A0A2J7QEY0 A0A0L8FGQ0 A0A1Y1K9I7 A0A1W4WAS5 W4XZ20 C3XT97 Q28DN4 K1QHN6 A7S4U2 T1J4E9 Q5U4A3 A0A1W2W4Z8 G7PN26 A0A2K6L8U9 A0A2K6P244 A0A2K6BTR6 G7NAZ7 A0A2K5KYP6 A0A096MNA7 A0A2K5ZHL7 H2P7E0 A0A0D9RIB8 G1QMF0 A0A2G8L2J7 A0A2K5J8N9 U4UPZ8 F7ACG4 A0A286XH06 K7BSJ4 G1U687 A7Z063 U4UT95 A0A2C9JEA9 N6U7W4 G3RVA7 I3MT98 G5B7J2 A0A3Q2H2I4 F7E3G3 A0A3P4PLA7 A0A0P7YEC5 F6QVX5 A0A1W5AZA3 A0A0P6JAG2 A0A3B3ST62 L9LCK4 G9KXK8 A0A2Y9KYL4 G3WP97 A0A2U3WS11 M3Z2V3 A0A2P6LBT2 A0A341AI29 A0A3Q7NWB8 M3X1I8 A0A3Q7ML03 A0A2Y9NJC0 A0A2Y9NU53 A0A341AGR2 A0A341AGQ5 A0A2Y9NNU5 Q6VEQ5 R7V9U1 F1SK83 A0A341AFV8 A0A287BQQ1 A0A2Y9NDJ0 A0A341ADS1 A0A2Y9NG41 A0A2Y9H4C6 A0A2U3YE77 A8K0Z3 A0A2U3YDX0 A0A3Q7T8B8 A0A1B0GJ14 E2QXP1 A0A3Q7RWZ3 G1LDI4 G1LDG8 A0A3Q7RZW7 A0A383Z564 A0A2G9RWY6 A0A3Q7WAA2 A0A384D8G4 A0A060WYS1 A0A383Z5G9 A0A383Z5D1
L7X0Q8 A0A194QGQ8 A0A212EP92 A0A0L7LB49 A0A067QUW1 A0A2J7QEZ0 A0A2J7QEY0 A0A0L8FGQ0 A0A1Y1K9I7 A0A1W4WAS5 W4XZ20 C3XT97 Q28DN4 K1QHN6 A7S4U2 T1J4E9 Q5U4A3 A0A1W2W4Z8 G7PN26 A0A2K6L8U9 A0A2K6P244 A0A2K6BTR6 G7NAZ7 A0A2K5KYP6 A0A096MNA7 A0A2K5ZHL7 H2P7E0 A0A0D9RIB8 G1QMF0 A0A2G8L2J7 A0A2K5J8N9 U4UPZ8 F7ACG4 A0A286XH06 K7BSJ4 G1U687 A7Z063 U4UT95 A0A2C9JEA9 N6U7W4 G3RVA7 I3MT98 G5B7J2 A0A3Q2H2I4 F7E3G3 A0A3P4PLA7 A0A0P7YEC5 F6QVX5 A0A1W5AZA3 A0A0P6JAG2 A0A3B3ST62 L9LCK4 G9KXK8 A0A2Y9KYL4 G3WP97 A0A2U3WS11 M3Z2V3 A0A2P6LBT2 A0A341AI29 A0A3Q7NWB8 M3X1I8 A0A3Q7ML03 A0A2Y9NJC0 A0A2Y9NU53 A0A341AGR2 A0A341AGQ5 A0A2Y9NNU5 Q6VEQ5 R7V9U1 F1SK83 A0A341AFV8 A0A287BQQ1 A0A2Y9NDJ0 A0A341ADS1 A0A2Y9NG41 A0A2Y9H4C6 A0A2U3YE77 A8K0Z3 A0A2U3YDX0 A0A3Q7T8B8 A0A1B0GJ14 E2QXP1 A0A3Q7RWZ3 G1LDI4 G1LDG8 A0A3Q7RZW7 A0A383Z564 A0A2G9RWY6 A0A3Q7WAA2 A0A384D8G4 A0A060WYS1 A0A383Z5G9 A0A383Z5D1
Ontologies
GO
GO:0071203
GO:0043014
GO:0005769
GO:0034314
GO:0003779
GO:0016197
GO:0055037
GO:0031901
GO:0042147
GO:0015031
GO:0055038
GO:0005829
GO:0006887
GO:0000145
GO:0071437
GO:0031274
GO:0031625
GO:0030335
GO:0022617
GO:0043231
GO:1990126
GO:0005814
GO:0005776
GO:0005770
GO:0046872
GO:0006801
GO:0030001
GO:0010507
GO:1904109
GO:0031396
GO:0031083
GO:0034383
GO:0043015
GO:0040038
GO:2000010
GO:0042098
GO:0099638
GO:0043553
GO:0002468
GO:0034394
GO:0001556
GO:0090306
GO:0005813
GO:0007032
GO:0050776
PANTHER
Topology
Subcellular location
Early endosome membrane
Recycling endosome membrane
Late endosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoplasmic vesicle Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Autophagosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoplasm Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoskeleton Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Microtubule organizing center Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Centrosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Centriole Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Recycling endosome membrane
Late endosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoplasmic vesicle Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Autophagosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoplasm Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Cytoskeleton Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Microtubule organizing center Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Centrosome Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
Centriole Localization to the endosome membrane is mediated via its interaction with WASHC2. With evidence from 1 publications.
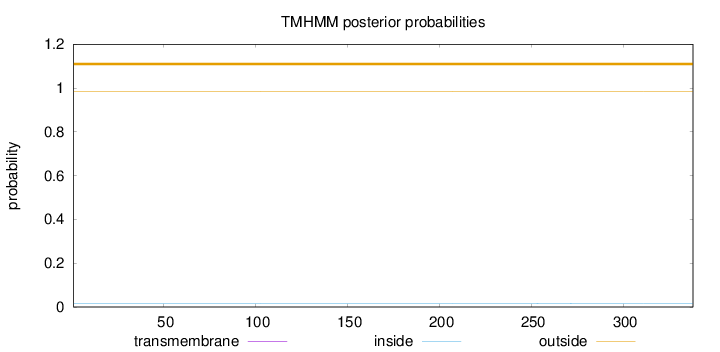
Length:
338
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00051
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01543
outside
1 - 338
Population Genetic Test Statistics
Pi
6.590049
Theta
11.261515
Tajima's D
-0.523821
CLR
0
CSRT
0.241637918104095
Interpretation
Uncertain
