Gene
KWMTBOMO16538 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005642
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_cytokine_receptor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.943
Sequence
CDS
ATGGAAAATCTTTCTGTGAATTTCGGAGGAACGAAGTGGAGTGATCTGCGGAGATTGCTTTTGTGTTTTAAATTAATGCTAATTTGGATTTTGTTGAGTTTGCCGTGTATATTCGCGCATTGTCAAGGCGTAGAAAATCTGTCAGTGGCAGTTTACCCCATGGGTAACCTGAAGGTGCACTATGGAGATTCCTTTGAACTATTTTGTGTGGCTGAAAAGTTCACGTCCGCCGATATTGAGTTTACAATGGGGGGTAAGACCATTCCTTCCGAAGTAGTAAACAGCACAACAATACGGCTTTACATGGAAAAACCCGAGAAACAAGTGAACACTTTTTATTGTCGTAATAAAAAAACAAACAAGATATGCACAAGCAGAGTGGTAGTGGACTCTACACCGATGGCAGTCACTGATTTTTCTTGTAAATCAAAAAATCTCGAGATTTTAAACTGTACTTGGATTGCTCCTGGACCTTTAAGTACGACCCAGAATTATATTACATTTTTGATTGATGGAAATGCTGTAGAACCCAGTACTCCGATCAGAGCGGATCAAACAAGATATTGGAACAGTTGGAATACTTACAGCATACCAAGATATAGACAAATGGAAAACAAGTATTATTTTTATTTGAGCTCGTGCAGTCCATTTGGTTGCAATAATCAAAGTTTTACAGTTGATCATTTCTCTATTGTGAAGCCTGATGCTCCAAGTAATTTGAAGGTAATAAGGAATTACACTAATGGGGTCTATCTACAATGGAATATACCTAATAACATGGTAGATTTATTGCCCTGTGGCGTTGATCATCGCATTGAATATCAAATTGATAAAATTGACAATAAATCTTATTTCCGAAGTGTGGATGCATCAAGTTTACCACCGAAAAATAGAACATATAAATTTTTACTAACAGATCTTCCTTATGCTCACATGAAATATGAAATCAGGATCTACATCAAATCTAAAGCTGCAGTGAAAGAAGAGTTTTGGTCAGACTTCACATATACCATGGTATATACACTTAGCGAGAGACCTAGAAGACCACCAGACACCGCAGTTGGAGCCTTTGATCAATCTTTATATCAAACAAATAGAATGATATACATTTATTGGAAACAACTTGAAGAATATGAAGAGGCAGGTACAAATTTCACTTACAAAGTTGTCGTTCATCATAAAAGCAATTATGAAACTTTGACAGCTGACAACAACAAAAGTTTGAGTTTTGTGGTCTTGGAAAAAGCACCATTTGATGCAATGGATGTCTACATTTGGTCTAGAAATGATAATGGATCTTCCATTAACAGCAGCCATCTGTACATTCCACCCAAAGAAGATACTGAAGCATTAAAAGTTTCTTCATTTACCAAACTAGCTTATGAAAATGGCGCATATAAATTGTCCTGGGTTGGTATAGAACATATTGACAATTATACACTTTTCTGGTGTCAACATAATAGTACAAAAATTTGTGCAGGCCGAATGAACTTCATTGTTTTAAGTGCCAATAAAACAATGCAAGTCATAGAACTACCTAGAGAAAACAGGTATCAGTTTGCCATATCGGCTAATAATGGGACGAAAACTAGTGGCATGGTATGGGCTATCTGTGATATATCTAAAGATGCAATTCCAATGTATGGTTTTCCGGTCAAAATGGACCATGGTATTCCAGGGAAAACTTTTGTAAAGATTAAATGGGCAATGACTTGTACACTTCAAGAAGGAATCATTACAGGATATTTGATCGCATATTGTCCTGCTCTAGATACCACTAATGTTTGTGCAACAAAAAATAAAACTAACTTATACATATCTAATCCTAAGCAAATGGAGATCAACATTACAAATCTTGAACCATACACTACATATATATTTACACTGGCATTGAACACCACTTATGGATTTAAAACAGTAGAAAATGCTTTCACGAGTATGACTACAACTGAAGATACTCCAACTAGTCCAGTTAATATTAATATAACCGATGTGCTTAGTGATTCATTGACGATTTCATGGGACCCACCTATTCATAAAAACGGAGTTATTGGAAAATATGTCATAAATAACTATGGTCAAGAAGTTTATGTAGAAAAAGTTTCAGATAAAGATGAAGATTCAAAAAGAAGGCATATTCGTTTGACTGGCTTACAGGGGTTTACTAATTATTCATTGAGTGTACAAGCATGTAGTACAGCTATAAGCAGTTGTTCGGTTTTGAATCCACATGATGCCCTTTTTGTCAGGACGAGAATAGGTCCTCCTAGTAAAATGAGGGCTCCAACTGTTAAAAATAATCCAGATTTTATTAAGTGGGTTCCACCAATAATACCAGGGGGGACGATTGACCTTTACCAAATTAAAAGGATTAAAGATGAGAGTGAAGCTGAAATAATAAATACTACAAACCTTTCATTTTCATTGATACATTGTGAAGGTGGAGTTGCCGGACAAACTTACCAAGTTAGAGCTGTTAATTTTGATGAAGATCCTTACCACGGTGCTTTAACTGACAATAAGGATGTATACCTACATAATGCTAACTATAAAGGTATACTAGAATATCCTGGAGAATGGAGTGAGCCAAGTTTTGTAAATTGTACAAGCAAAGAACATGTGACACTAACTCTTATTTTAATGGGAATATTTATGATAATTGGTATTATGTATGGTTTTATTAAATGTTACAAGAAATACCGAAAAATGGAAGATATTAAACCAGTTTTACCAAGTGGACTAGGAGTCCCCGAAAAGGATATTTCGAAATATGCTATTGGTGGTTGGAATCCTACAAACAAGGATGAGAAACCATCTTCAGACGAAATGTTGCTATTACCAAATAGTAGAACCACTGTATCTTCAGAATTGAAACAGAAAGAAAACAATAACAGTGCTTCTAGTGATCACACTGATAGTACAGCTTTATCAGATACATCACATGGGCCTGTAGAAAGACAAATCTCCTCATCTGATGATGGATCTAATTCATCACTACATCTTGAAGTAGAACCTACGAGAACAGCTGACAGTAACATCGATGGCGAATCATGCAATTCCGAAACTGAAACATCACAAATACCGTCACCATTCTTTAATGATAATACTTTCAAGAAAAATCCATGTGGCTATGTGCAACAATCTGTAGTGAGTCCTAATACAGGATATGTTCAAAGTGTTCAAGCAGGCGTTACAAACCCACCACAACCCTCATCACTCAGTGCCAGCTCTAGTTATGTAATGGCAGCTCTGTCTCCACCTATATTTACGACACGTGTTGCTCAACCTTGCGTAACGAGTTCACAATCAGCAGGAGGCTATGTGCACCCTGAAGGTGCTCAAACTATATCAGCAATGAATTTCCCGAAATTGGGGCAGACAACGAACAAACTGTTTGGTCCTGAAAGCTTGCCAACAATGCAAACCTTGCCACCAGCGAAACACACTGCAGACAGCAGCTACATTCAACTGCAATCTCTAGATTCCTTGCCCAGACACAAACATCCTGCCCGAAGTACAGTGCCATTAAAGACGCCAGCGTCTACCGGCTACGTCCGCCAAGGTGACGCTGTTATTAATAAGCACCTGAATAATATGCTATCAGGTGTTCATTGTGGCGAAGAATCGGCTATTTTGGATCCTGCAATGTCGCCTGACGCGTACTGCAGATTTTCATGGAGTACCGATCCAGCCAATGACAACATTCATTCGTTACTCGCCGATTCGCCTAGATTAAATTCATCGAAAAACGGAGTCAATCATTAG
Protein
MENLSVNFGGTKWSDLRRLLLCFKLMLIWILLSLPCIFAHCQGVENLSVAVYPMGNLKVHYGDSFELFCVAEKFTSADIEFTMGGKTIPSEVVNSTTIRLYMEKPEKQVNTFYCRNKKTNKICTSRVVVDSTPMAVTDFSCKSKNLEILNCTWIAPGPLSTTQNYITFLIDGNAVEPSTPIRADQTRYWNSWNTYSIPRYRQMENKYYFYLSSCSPFGCNNQSFTVDHFSIVKPDAPSNLKVIRNYTNGVYLQWNIPNNMVDLLPCGVDHRIEYQIDKIDNKSYFRSVDASSLPPKNRTYKFLLTDLPYAHMKYEIRIYIKSKAAVKEEFWSDFTYTMVYTLSERPRRPPDTAVGAFDQSLYQTNRMIYIYWKQLEEYEEAGTNFTYKVVVHHKSNYETLTADNNKSLSFVVLEKAPFDAMDVYIWSRNDNGSSINSSHLYIPPKEDTEALKVSSFTKLAYENGAYKLSWVGIEHIDNYTLFWCQHNSTKICAGRMNFIVLSANKTMQVIELPRENRYQFAISANNGTKTSGMVWAICDISKDAIPMYGFPVKMDHGIPGKTFVKIKWAMTCTLQEGIITGYLIAYCPALDTTNVCATKNKTNLYISNPKQMEINITNLEPYTTYIFTLALNTTYGFKTVENAFTSMTTTEDTPTSPVNINITDVLSDSLTISWDPPIHKNGVIGKYVINNYGQEVYVEKVSDKDEDSKRRHIRLTGLQGFTNYSLSVQACSTAISSCSVLNPHDALFVRTRIGPPSKMRAPTVKNNPDFIKWVPPIIPGGTIDLYQIKRIKDESEAEIINTTNLSFSLIHCEGGVAGQTYQVRAVNFDEDPYHGALTDNKDVYLHNANYKGILEYPGEWSEPSFVNCTSKEHVTLTLILMGIFMIIGIMYGFIKCYKKYRKMEDIKPVLPSGLGVPEKDISKYAIGGWNPTNKDEKPSSDEMLLLPNSRTTVSSELKQKENNNSASSDHTDSTALSDTSHGPVERQISSSDDGSNSSLHLEVEPTRTADSNIDGESCNSETETSQIPSPFFNDNTFKKNPCGYVQQSVVSPNTGYVQSVQAGVTNPPQPSSLSASSSYVMAALSPPIFTTRVAQPCVTSSQSAGGYVHPEGAQTISAMNFPKLGQTTNKLFGPESLPTMQTLPPAKHTADSSYIQLQSLDSLPRHKHPARSTVPLKTPASTGYVRQGDAVINKHLNNMLSGVHCGEESAILDPAMSPDAYCRFSWSTDPANDNIHSLLADSPRLNSSKNGVNH
Summary
Uniprot
EMBL
BABH01020206
NWSH01001832
PCG69962.1
ODYU01006205
SOQ47849.1
KQ461155
+ More
KPJ08303.1 KQ459232 KPJ02630.1 FP102340 CBH09280.1 GDQN01001839 JAT89215.1 AGBW02013517 OWR43293.1 KK853224 KDR09708.1 NEVH01002541 PNF42340.1 GECZ01020155 JAS49614.1 KQ414617 KOC68210.1 GBYB01005083 GBYB01005085 JAG74850.1 JAG74852.1 ADTU01024298 KK107274 QOIP01000008 EZA53737.1 RLU19892.1 KQ976424 KYM88588.1 KQ978983 KYN26832.1 LJIG01009181 KRT83594.1 KZ288291 PBC29205.1 GGMR01001923 MBY14542.1 DS235405 EEB15556.1 GFXV01002716 MBW14521.1 JH432010 DS232723 EDS45159.1 GDIQ01035546 JAN59191.1 GDIQ01035545 JAN59192.1
KPJ08303.1 KQ459232 KPJ02630.1 FP102340 CBH09280.1 GDQN01001839 JAT89215.1 AGBW02013517 OWR43293.1 KK853224 KDR09708.1 NEVH01002541 PNF42340.1 GECZ01020155 JAS49614.1 KQ414617 KOC68210.1 GBYB01005083 GBYB01005085 JAG74850.1 JAG74852.1 ADTU01024298 KK107274 QOIP01000008 EZA53737.1 RLU19892.1 KQ976424 KYM88588.1 KQ978983 KYN26832.1 LJIG01009181 KRT83594.1 KZ288291 PBC29205.1 GGMR01001923 MBY14542.1 DS235405 EEB15556.1 GFXV01002716 MBW14521.1 JH432010 DS232723 EDS45159.1 GDIQ01035546 JAN59191.1 GDIQ01035545 JAN59192.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
6IAA
E-value=0.00508117,
Score=99
Ontologies
GO
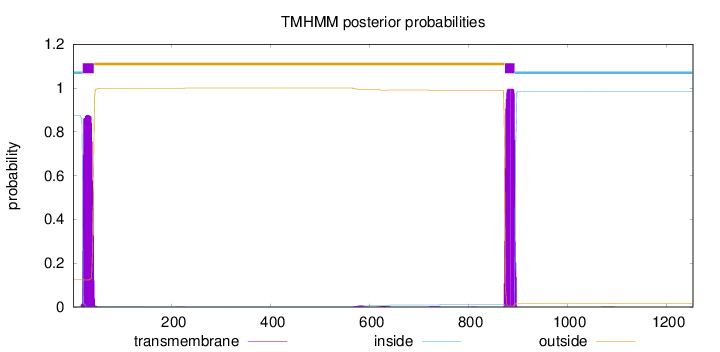
Topology
Length:
1254
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.4895400000001
Exp number, first 60 AAs:
19.01989
Total prob of N-in:
0.87579
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 873
TMhelix
874 - 893
inside
894 - 1254
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.000008
CSRT
0
Interpretation
Uncertain
