Gene
KWMTBOMO16537
Pre Gene Modal
BGIBMGA005562
Annotation
PREDICTED:_uncharacterized_protein_KIAA0195?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.05
Sequence
CDS
ATGGATGATAACAACGAAGAAATATTAGGCTATTCAACTAGACATGCTCTGATCAAATTACGCAACGACATTAGCGGCGTGATTGAAGAATATAAAAAGAAGATTCCCACTGGATTTCGCTTGGCACGTGATATTATTTCGTTTTCATCAGAGAAGGTTCTACTTTCGGGTGTTGCATACACAATTACATTGATTTACATAGTATGCCTATTATTAACATACCTATTTGGGGTCTCAGTACAGGCCCATGCATATCTGTTATGGGAGGCTTTTTTTCTATTAATCATATTAATAATTAACTTTTTGGTGGCATTTAACGAAGAATATTTATATCAAAATGAAATACCTCATAGAGTAAAAAAAGTGCTGGAAACAATGACTAACGCTATCAATAGAGTGAAATGGAAAGAAGAATATTATCCTCACCTATGTGCACCATATTCGCCGTGTGTAACTTTACAATGGACATACAGAGATGGTAACAAGATTTATTGA
Protein
MDDNNEEILGYSTRHALIKLRNDISGVIEEYKKKIPTGFRLARDIISFSSEKVLLSGVAYTITLIYIVCLLLTYLFGVSVQAHAYLLWEAFFLLIILIINFLVAFNEEYLYQNEIPHRVKKVLETMTNAINRVKWKEEYYPHLCAPYSPCVTLQWTYRDGNKIY
Summary
Uniprot
H9J7S0
A0A2A4JEV8
A0A1E1WM40
A0A2H1W431
A0A3S2L8H5
A0A194QSL0
+ More
D0AB86 L7X3W0 A0A212EPB0 A0A194QB23 A0A1E1WHE4 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9RMK5 A0A0C9PL89 A0A0N0U4H9 A0A087ZXX4 N6UJ90 E2B474 A0A0A9XU67 U4UKU9 A0A2R7VPD4 A0A195B5D9 A0A151J7P1 A0A158NM24 A0A2P8YEJ8 A0A151JXP4 A0A146LYF4 A0A146M4X0 A0A146LUX9 F4WHR9 A0A2A3EAG4 A0A067QXM3 A0A023F1Q2 A0A310SMW2 D6WX73 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DHS1 A0A1B6BXT8 A0A1B6DN27 A0A1B6DZA4 A0A1B6CFK5 A0A1B6D8B3 A0A1B6EF44 A0A1B6CRC8 A0A1B6C8E3 A0A1B6DTF3 V9IFN6 A0A0L7QQV6 A0A026WG49 A0A3L8DJS8 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1Y1JTV9 A0A1Y1JWC3 A0A232ER02 A0A1L8D957 A0A1D2NIX0 A0A1B0CT49 A0A2M4AB14 A0A0N8ES84 A0A182FJZ0 A0A084VAA9 A0A1B6HB37 A0A1B6JB83 A0A182LXB7 A0A1B6HLI4 A0A1B6JBX6 A0A182RV48 A0A2M4CP22 W5JKG4 A0A182PQ59 A0A034V6X9 A0A034VAE7 A0A0K8VTX2 A0A182JHX8 A0A0K8UU51 A0A0K8TVT7 A0A0K8U2V1 A0A0K8UN70 A0A0K8WCQ8 A0A182H3K9 A0A182GSD3 W8AHZ9 A0A1B6KT90 W8B4C2 A0A0A1XMR0 A0A182QD36 A0A0A1X921 A0A182W4Q9
D0AB86 L7X3W0 A0A212EPB0 A0A194QB23 A0A1E1WHE4 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9RMK5 A0A0C9PL89 A0A0N0U4H9 A0A087ZXX4 N6UJ90 E2B474 A0A0A9XU67 U4UKU9 A0A2R7VPD4 A0A195B5D9 A0A151J7P1 A0A158NM24 A0A2P8YEJ8 A0A151JXP4 A0A146LYF4 A0A146M4X0 A0A146LUX9 F4WHR9 A0A2A3EAG4 A0A067QXM3 A0A023F1Q2 A0A310SMW2 D6WX73 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DHS1 A0A1B6BXT8 A0A1B6DN27 A0A1B6DZA4 A0A1B6CFK5 A0A1B6D8B3 A0A1B6EF44 A0A1B6CRC8 A0A1B6C8E3 A0A1B6DTF3 V9IFN6 A0A0L7QQV6 A0A026WG49 A0A3L8DJS8 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1Y1JTV9 A0A1Y1JWC3 A0A232ER02 A0A1L8D957 A0A1D2NIX0 A0A1B0CT49 A0A2M4AB14 A0A0N8ES84 A0A182FJZ0 A0A084VAA9 A0A1B6HB37 A0A1B6JB83 A0A182LXB7 A0A1B6HLI4 A0A1B6JBX6 A0A182RV48 A0A2M4CP22 W5JKG4 A0A182PQ59 A0A034V6X9 A0A034VAE7 A0A0K8VTX2 A0A182JHX8 A0A0K8UU51 A0A0K8TVT7 A0A0K8U2V1 A0A0K8UN70 A0A0K8WCQ8 A0A182H3K9 A0A182GSD3 W8AHZ9 A0A1B6KT90 W8B4C2 A0A0A1XMR0 A0A182QD36 A0A0A1X921 A0A182W4Q9
Pubmed
EMBL
BABH01020203
BABH01020204
BABH01020205
BABH01020206
NWSH01001832
PCG69963.1
+ More
GDQN01002961 JAT88093.1 ODYU01006205 SOQ47850.1 RSAL01000090 RVE48102.1 KQ461155 KPJ08304.1 FP102340 CBH09281.1 KC469893 AGC92690.2 AGBW02013517 OWR43291.1 KQ459232 KPJ02629.1 GDQN01004645 JAT86409.1 GBYB01009583 JAG79350.1 GBYB01011032 JAG80799.1 GBYB01001767 JAG71534.1 GBYB01009580 JAG79347.1 GBYB01001768 JAG71535.1 KQ435834 KOX71551.1 APGK01022082 KB740398 ENN80731.1 GL445515 EFN89491.1 GBHO01045649 GBHO01026606 GBHO01021166 JAF97954.1 JAG16998.1 JAG22438.1 KB632255 ERL90630.1 KK854009 PTY09219.1 KQ976604 KYM79404.1 KQ979658 KYN19892.1 ADTU01020204 PYGN01000660 PSN42666.1 KQ981561 KYN39827.1 GDHC01006927 JAQ11702.1 GDHC01003801 JAQ14828.1 GDHC01007108 JAQ11521.1 GL888167 EGI66276.1 KZ288308 PBC28715.1 KK853134 KDR10862.1 GBBI01003783 JAC14929.1 KQ759847 OAD62585.1 KQ971361 EFA08800.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01026348 JAS10950.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 GEDC01006317 JAS30981.1 GEDC01025148 JAS12150.1 GEDC01015412 JAS21886.1 GEDC01000814 JAS36484.1 GEDC01024062 GEDC01021433 JAS13236.1 JAS15865.1 GEDC01027521 JAS09777.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 JR040354 AEY59099.1 KQ414784 KOC61013.1 KK107238 EZA54621.1 QOIP01000007 RLU20670.1 GEZM01101564 GEZM01101559 GEZM01101557 JAV52545.1 GEZM01101544 JAV52589.1 GEZM01101577 GEZM01101572 GEZM01101571 GEZM01101563 GEZM01101562 GEZM01101561 GEZM01101555 GEZM01101554 GEZM01101552 GEZM01101543 GEZM01101542 JAV52531.1 GEZM01101574 JAV52505.1 GEZM01101579 GEZM01101578 GEZM01101575 GEZM01101568 GEZM01101560 GEZM01101558 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101541 GEZM01101540 JAV52604.1 GEZM01101545 JAV52588.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 NNAY01002685 OXU20778.1 GFDF01011093 JAV02991.1 LJIJ01000028 ODN05210.1 AJWK01027094 GGFK01004662 MBW37983.1 GDUN01000178 JAN95741.1 ATLV01003052 KE524109 KFB34903.1 GECU01035785 JAS71921.1 GECU01011423 JAS96283.1 AXCM01001247 GECU01032144 JAS75562.1 GECU01011041 JAS96665.1 GGFL01002892 MBW67070.1 ADMH02000938 ETN64626.1 GAKP01019871 JAC39081.1 GAKP01019870 JAC39082.1 GDHF01009982 JAI42332.1 AXCP01007583 GDHF01022193 JAI30121.1 GDHF01033916 JAI18398.1 GDHF01031513 JAI20801.1 GDHF01024534 GDHF01015083 JAI27780.1 JAI37231.1 GDHF01003346 JAI48968.1 JXUM01107644 JXUM01107645 JXUM01107646 KQ565160 KXJ71272.1 JXUM01084398 KQ563435 KXJ73833.1 GAMC01018440 JAB88115.1 GEBQ01025356 JAT14621.1 GAMC01018439 JAB88116.1 GBXI01002050 JAD12242.1 AXCN02001394 GBXI01006665 JAD07627.1
GDQN01002961 JAT88093.1 ODYU01006205 SOQ47850.1 RSAL01000090 RVE48102.1 KQ461155 KPJ08304.1 FP102340 CBH09281.1 KC469893 AGC92690.2 AGBW02013517 OWR43291.1 KQ459232 KPJ02629.1 GDQN01004645 JAT86409.1 GBYB01009583 JAG79350.1 GBYB01011032 JAG80799.1 GBYB01001767 JAG71534.1 GBYB01009580 JAG79347.1 GBYB01001768 JAG71535.1 KQ435834 KOX71551.1 APGK01022082 KB740398 ENN80731.1 GL445515 EFN89491.1 GBHO01045649 GBHO01026606 GBHO01021166 JAF97954.1 JAG16998.1 JAG22438.1 KB632255 ERL90630.1 KK854009 PTY09219.1 KQ976604 KYM79404.1 KQ979658 KYN19892.1 ADTU01020204 PYGN01000660 PSN42666.1 KQ981561 KYN39827.1 GDHC01006927 JAQ11702.1 GDHC01003801 JAQ14828.1 GDHC01007108 JAQ11521.1 GL888167 EGI66276.1 KZ288308 PBC28715.1 KK853134 KDR10862.1 GBBI01003783 JAC14929.1 KQ759847 OAD62585.1 KQ971361 EFA08800.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01026348 JAS10950.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 GEDC01006317 JAS30981.1 GEDC01025148 JAS12150.1 GEDC01015412 JAS21886.1 GEDC01000814 JAS36484.1 GEDC01024062 GEDC01021433 JAS13236.1 JAS15865.1 GEDC01027521 JAS09777.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 JR040354 AEY59099.1 KQ414784 KOC61013.1 KK107238 EZA54621.1 QOIP01000007 RLU20670.1 GEZM01101564 GEZM01101559 GEZM01101557 JAV52545.1 GEZM01101544 JAV52589.1 GEZM01101577 GEZM01101572 GEZM01101571 GEZM01101563 GEZM01101562 GEZM01101561 GEZM01101555 GEZM01101554 GEZM01101552 GEZM01101543 GEZM01101542 JAV52531.1 GEZM01101574 JAV52505.1 GEZM01101579 GEZM01101578 GEZM01101575 GEZM01101568 GEZM01101560 GEZM01101558 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101541 GEZM01101540 JAV52604.1 GEZM01101545 JAV52588.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 NNAY01002685 OXU20778.1 GFDF01011093 JAV02991.1 LJIJ01000028 ODN05210.1 AJWK01027094 GGFK01004662 MBW37983.1 GDUN01000178 JAN95741.1 ATLV01003052 KE524109 KFB34903.1 GECU01035785 JAS71921.1 GECU01011423 JAS96283.1 AXCM01001247 GECU01032144 JAS75562.1 GECU01011041 JAS96665.1 GGFL01002892 MBW67070.1 ADMH02000938 ETN64626.1 GAKP01019871 JAC39081.1 GAKP01019870 JAC39082.1 GDHF01009982 JAI42332.1 AXCP01007583 GDHF01022193 JAI30121.1 GDHF01033916 JAI18398.1 GDHF01031513 JAI20801.1 GDHF01024534 GDHF01015083 JAI27780.1 JAI37231.1 GDHF01003346 JAI48968.1 JXUM01107644 JXUM01107645 JXUM01107646 KQ565160 KXJ71272.1 JXUM01084398 KQ563435 KXJ73833.1 GAMC01018440 JAB88115.1 GEBQ01025356 JAT14621.1 GAMC01018439 JAB88116.1 GBXI01002050 JAD12242.1 AXCN02001394 GBXI01006665 JAD07627.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000053105 UP000005203 UP000019118 UP000008237 UP000030742 UP000078540 UP000078492 UP000005205 UP000245037 UP000078541 UP000007755 UP000242457 UP000027135 UP000007266 UP000053825 UP000053097 UP000279307 UP000215335 UP000094527 UP000092461 UP000069272 UP000030765 UP000075883 UP000075900 UP000000673 UP000075885 UP000075880 UP000069940 UP000249989 UP000075886 UP000075920
UP000053105 UP000005203 UP000019118 UP000008237 UP000030742 UP000078540 UP000078492 UP000005205 UP000245037 UP000078541 UP000007755 UP000242457 UP000027135 UP000007266 UP000053825 UP000053097 UP000279307 UP000215335 UP000094527 UP000092461 UP000069272 UP000030765 UP000075883 UP000075900 UP000000673 UP000075885 UP000075880 UP000069940 UP000249989 UP000075886 UP000075920
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J7S0
A0A2A4JEV8
A0A1E1WM40
A0A2H1W431
A0A3S2L8H5
A0A194QSL0
+ More
D0AB86 L7X3W0 A0A212EPB0 A0A194QB23 A0A1E1WHE4 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9RMK5 A0A0C9PL89 A0A0N0U4H9 A0A087ZXX4 N6UJ90 E2B474 A0A0A9XU67 U4UKU9 A0A2R7VPD4 A0A195B5D9 A0A151J7P1 A0A158NM24 A0A2P8YEJ8 A0A151JXP4 A0A146LYF4 A0A146M4X0 A0A146LUX9 F4WHR9 A0A2A3EAG4 A0A067QXM3 A0A023F1Q2 A0A310SMW2 D6WX73 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DHS1 A0A1B6BXT8 A0A1B6DN27 A0A1B6DZA4 A0A1B6CFK5 A0A1B6D8B3 A0A1B6EF44 A0A1B6CRC8 A0A1B6C8E3 A0A1B6DTF3 V9IFN6 A0A0L7QQV6 A0A026WG49 A0A3L8DJS8 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1Y1JTV9 A0A1Y1JWC3 A0A232ER02 A0A1L8D957 A0A1D2NIX0 A0A1B0CT49 A0A2M4AB14 A0A0N8ES84 A0A182FJZ0 A0A084VAA9 A0A1B6HB37 A0A1B6JB83 A0A182LXB7 A0A1B6HLI4 A0A1B6JBX6 A0A182RV48 A0A2M4CP22 W5JKG4 A0A182PQ59 A0A034V6X9 A0A034VAE7 A0A0K8VTX2 A0A182JHX8 A0A0K8UU51 A0A0K8TVT7 A0A0K8U2V1 A0A0K8UN70 A0A0K8WCQ8 A0A182H3K9 A0A182GSD3 W8AHZ9 A0A1B6KT90 W8B4C2 A0A0A1XMR0 A0A182QD36 A0A0A1X921 A0A182W4Q9
D0AB86 L7X3W0 A0A212EPB0 A0A194QB23 A0A1E1WHE4 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9RMK5 A0A0C9PL89 A0A0N0U4H9 A0A087ZXX4 N6UJ90 E2B474 A0A0A9XU67 U4UKU9 A0A2R7VPD4 A0A195B5D9 A0A151J7P1 A0A158NM24 A0A2P8YEJ8 A0A151JXP4 A0A146LYF4 A0A146M4X0 A0A146LUX9 F4WHR9 A0A2A3EAG4 A0A067QXM3 A0A023F1Q2 A0A310SMW2 D6WX73 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DHS1 A0A1B6BXT8 A0A1B6DN27 A0A1B6DZA4 A0A1B6CFK5 A0A1B6D8B3 A0A1B6EF44 A0A1B6CRC8 A0A1B6C8E3 A0A1B6DTF3 V9IFN6 A0A0L7QQV6 A0A026WG49 A0A3L8DJS8 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1Y1JTV9 A0A1Y1JWC3 A0A232ER02 A0A1L8D957 A0A1D2NIX0 A0A1B0CT49 A0A2M4AB14 A0A0N8ES84 A0A182FJZ0 A0A084VAA9 A0A1B6HB37 A0A1B6JB83 A0A182LXB7 A0A1B6HLI4 A0A1B6JBX6 A0A182RV48 A0A2M4CP22 W5JKG4 A0A182PQ59 A0A034V6X9 A0A034VAE7 A0A0K8VTX2 A0A182JHX8 A0A0K8UU51 A0A0K8TVT7 A0A0K8U2V1 A0A0K8UN70 A0A0K8WCQ8 A0A182H3K9 A0A182GSD3 W8AHZ9 A0A1B6KT90 W8B4C2 A0A0A1XMR0 A0A182QD36 A0A0A1X921 A0A182W4Q9
Ontologies
PANTHER
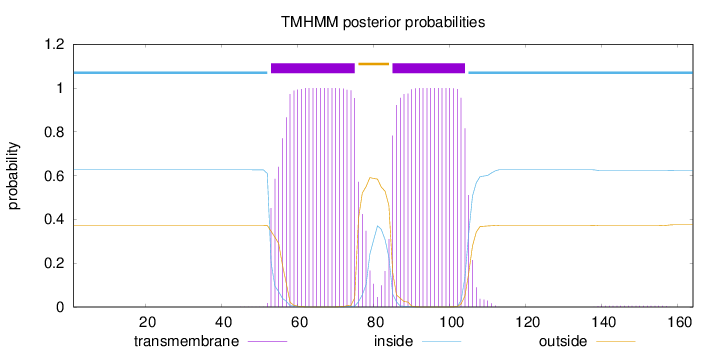
Topology
Length:
164
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.84717
Exp number, first 60 AAs:
6.28949
Total prob of N-in:
0.62702
inside
1 - 52
TMhelix
53 - 75
outside
76 - 84
TMhelix
85 - 104
inside
105 - 164
Population Genetic Test Statistics
Pi
3.425465
Theta
35.131031
Tajima's D
-0.773334
CLR
0
CSRT
0.179791010449478
Interpretation
Uncertain
