Gene
KWMTBOMO16536
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.713
Sequence
CDS
ATGCCGATTGGGAAAGTTGAAAGTTTTGATATGGGATCTCAAAATTGGGACACATACATTCGTCGTGTAAAACAATTCATGGAACTCAACGACATTGAACAGAGACTTCACGTTGCAACGCTCGTGACCTTAGTTGGGTCTGAGTGCTATGACTTAATGTGTGACTTGTGTGCTCCGGACTTACCAGAAGAAAAGGATTTCGACATACTCGTAAAATTAGTCAAAGATCATCTAGAACCTGAAAGATCGGAAATAGCAGAACGTCATATCTTCAGGCAAAGAAAGCAGCAAAATAACGAAAGCATTCGTTCCTACCTACAAAGTTTGAAACATTTGGCTAAAACTTGCAATTTCGGCATCACATTAGAAATTAATCTACGCGATCAATTCGTATCAGGATTTTATAGTGAGGAGATGCGATCAAGGCTGTTTGCTGAAAAAGATATAGATTACAAGCGCGCTGTTGAGTTGGCATCAGCACTGGAAGCAGCGGATCGACATGCGATGGCGGCTGGGGGCATTGGGGGCAGCTCACAGGCTGCGTCCGAGAGCGAAGGCATGCATCGGGTCGGCGGAGCGCGCGCGGAGGCTCAATGCGCCTGCAGACGCTGCGGCAAGCTGGGCCATGCGGAGGGTCGGTGTCGGTACAAGCACTACACCTGTGACTCATGTGGCGAGAAGGGACACCTAAAATCAATGTGTCGGAACCGTAAAGGTAACCAAAAGATTGAAAAAAACCGAGGTCAGTACTATTTAAACCATAGTGACTGTGATTCTAGTGATAATAGTTTTTATAACTTAAAGGACCTGATCAGTAGCGAGGGTGAAGGACCATATTACGTGAAAATTTTTGTACAAAATGTTCCATGTAAATTTGAGATTGATACGGGTTGTAAAATTTCGGCCATATCTAAAGCTTTTTATGAAAAGCATTTTATGTGTATTCCTATACAAAGCGAAATCTTGAATTTTAGTTCATATACAGGCGATGTAATTGAGACTATCGGTTATATAATGGTAGACGTATCTTGTGGTCGTGAGTCCGCCAAATTAAAACTATATGTAATTAAAAACGGCGGCCCGCCACTGCTAGGCCGAATTTGGATTAAAATATTAAAATTAAGTTTTCTAAAATGTCATTCGATTTCTGAGACGACCTCGACGGTGGAGGCTTTACGAAACGAATTTCCAGAAGTGTTTGCTGGAGGTTTAGGAACATGTAAATCACGCATAAAATTACATCTAAAAGAAAAGACGCCAATATTTGTAAAAGCGCGGGCGCTGCCACTGGCGTTGCGCGAGCGCGTTGAGCGAGAACTTGATCGTTTACAAAGTGAGGGAGTTATTTACAAAGTAGATAGGTCTGATTACGGCACGCCTATCGTACCAGTCGTTAAGGCAAATGGCGATATTCGTATTTGCGGCGACTATAAAATCACAATAAACCCACGTCTGAAGGATTATCACTATCCTTTGCCGCACATTAAAGATTTGTTTGCCACTTTAGGTGGTGGTGAATATTACACAAAATTGGATTTATCTAATGCATTCCAGCAATGCCTCTTAGACGAAGAATCCCAGCCGATGACAGCGATCACGACACACATTGGTACTTTTGTCTATAAGCGCGTTCCGTTTGGAATTAAATGCATTCCGGAAAACTTTCAGAAGATTATGGAAGAGATTCTAAGTGGTCTGCCGTCAACCGTTATTTTTGCGGACGATATATGTGTAACCGGAAAAAACAGATTTGAACATTTATCAAATCTACGAGCAGTGTTGAGGAGACTCAAAGAAAACGGATTAAGAATTAATTATGACAAATGTAAATTCTTTCAAAGTTCTGTTACGTATTTAGGCTATAAAATAGACAAACAGGGCTTACACACTGACAAAAAGAAAGTTGATGCTATAGTGGCCGCCCCCGCACCTACGAATTTAACTCAATTGAGAAGTTTTATGGGTCTTGTTAATTTTTATTCGAACTTCGTTGTTAATATGAGTGATATTTTGAAACCTTTGTATAATTTGTTAAAAAAAAATGTAGAATGGTGTTGGACAGATAAATGTCATCAAGCATTTATACGAATTAAAAAAGTGTTAAGCAGCTCTCCGGTACTGGCACATTATAACCCAAATTCGCCATTAGTGCTGTCCGTTGACAGCAGCCAGTACGGACTGGGCGCCGTCCTCACTCAGAAGCACATCGACGGCACTGAGCGGCTCGTCAGCTGTGGCTCACGGACTCTCGCGCCAGCCGAACTTAACTATTCTCAGCTGGACAAAGAAGCTTTGGCTATAATGTTTGGGGTTACTAAACACCATCAGTATTTGTACGGGAGACGTTTTGTTTTACGAAGTGATCATAAGGCTTTGAGTTATATTCTTGGAAAGAATAAAGGTATACCTCTAACCGCCGCGAGTCGTCTGCAACGATTTGCTGTAAAGCTAGCTGCGTATGATTTTGAGATCGAATTTATTAGTTCTACAGATAATTGCCAGGCTGACGCGCTATCACGACTTCCACTGGATACTAATTTTAAGAATAATATTAAGGAATATAATTACCTAAACTTCGTTCAAGAAAGCTTTCCACTCAGTTTTAAAGAAATTAAAAAGGAAGTTGCGAAGGATCCTTTACTAAACAAAATTTACGGTTACGTCATGTTTGGCTGGCCCTCTGTCAGTAAAGAACAAGCAGAAAAACCCTATTTTAACAGAAGAGAATCCATACATATTGATCACGGTTGTCTTGTATGGGGTTACAGAATAATAATTCCAACTTCATTGCGAGAGACTGTGTTACAGGAGTTACATGAGGGACATCCGGGTATTGTTAAGATGAAACAATTAAGCCGGAACTACGTATGGTGGGAGACAGTCGATGCGGACATTGAACGTGTATGTGCTCAGTGCCAGGCCTGTCGATCTCAGCGCGCTGCACCACCATCGGTGCCGCTGCACTCGTGGCCATGGCCGGAAGAGCCGTGGGCGCGCCTTAATCTTGATTTCTTGGGACCGTTCAATAACAAATATTACTTAATTATAGTGGACGCTCATTCAAAATGGATCGAGGTGGAAAAATTGAGCTGTACATCAGCCACGGCTGTAATCGCATGTCTCAGACGATTGTTTGCTCGATTTGGCTTAGCAAAAAGGGTAGTTACTGATAACGGACCACCTTTCTCGTCGGTCGAATTTAGATCATATTTAAGTAAAAACGGTATAAAACATTTACCGGTGGCTCCGTATCATCCATCTAGCAATGGTGCAGCGGAAAACGCCGTAAAAACCATAAAAAGAGTGTTGAAAAAGGCTTTAATTGAAAAGGAAGACGACAATACCGCCTTGACAAAATTTCTCTTTATGTACAGAAATTCTGAGCATAGCACTACAGGGCGCGAACCAGCAGTTGCTTTGTTCGGACGTCGCCTGCGTGGAAGACTGGATCTGTTGCGTCCCAATACCAGCTCCATAGTACGAGAAGCACAGCTGGCAAGCGAAAGCTATAATGCAGGCACAGCCGGATATAGGGAGGCTGCAGAAGGAGACTCGGTCCTATTGCAAGATTTCACACAGGGAAAAAAGAAATGGACCCCAGGAACAATAAAAAACCGCTCAGGTCCGGTGACATACAGGGTAACAGCAGATGACGGGCGGGTACACAAACGTCACATCGACCAGATTTTAATAAGTAATAATAGTCGAAAATCACGTTATTCTTTGTCCAAATTAAACAATGATTTCGAGACAAAATCAGTAAATGATAGTGTGGGTGAAGCTCGGGAAAGTCCGGGTATGACCGGAGAAAAGGAGCTCAACTCCAGCTATGACACGGGACCGGAAAGTCCACAACCTGTGACTCCTAGTCACTCGCCGGCGCCGAATGTCTTGCAACCATCGCGTCAGCGCCGTAAAGCTGCTTTAACGTGTATTCAGAGAATTAAGGACCAAAATTACTAA
Protein
MPIGKVESFDMGSQNWDTYIRRVKQFMELNDIEQRLHVATLVTLVGSECYDLMCDLCAPDLPEEKDFDILVKLVKDHLEPERSEIAERHIFRQRKQQNNESIRSYLQSLKHLAKTCNFGITLEINLRDQFVSGFYSEEMRSRLFAEKDIDYKRAVELASALEAADRHAMAAGGIGGSSQAASESEGMHRVGGARAEAQCACRRCGKLGHAEGRCRYKHYTCDSCGEKGHLKSMCRNRKGNQKIEKNRGQYYLNHSDCDSSDNSFYNLKDLISSEGEGPYYVKIFVQNVPCKFEIDTGCKISAISKAFYEKHFMCIPIQSEILNFSSYTGDVIETIGYIMVDVSCGRESAKLKLYVIKNGGPPLLGRIWIKILKLSFLKCHSISETTSTVEALRNEFPEVFAGGLGTCKSRIKLHLKEKTPIFVKARALPLALRERVERELDRLQSEGVIYKVDRSDYGTPIVPVVKANGDIRICGDYKITINPRLKDYHYPLPHIKDLFATLGGGEYYTKLDLSNAFQQCLLDEESQPMTAITTHIGTFVYKRVPFGIKCIPENFQKIMEEILSGLPSTVIFADDICVTGKNRFEHLSNLRAVLRRLKENGLRINYDKCKFFQSSVTYLGYKIDKQGLHTDKKKVDAIVAAPAPTNLTQLRSFMGLVNFYSNFVVNMSDILKPLYNLLKKNVEWCWTDKCHQAFIRIKKVLSSSPVLAHYNPNSPLVLSVDSSQYGLGAVLTQKHIDGTERLVSCGSRTLAPAELNYSQLDKEALAIMFGVTKHHQYLYGRRFVLRSDHKALSYILGKNKGIPLTAASRLQRFAVKLAAYDFEIEFISSTDNCQADALSRLPLDTNFKNNIKEYNYLNFVQESFPLSFKEIKKEVAKDPLLNKIYGYVMFGWPSVSKEQAEKPYFNRRESIHIDHGCLVWGYRIIIPTSLRETVLQELHEGHPGIVKMKQLSRNYVWWETVDADIERVCAQCQACRSQRAAPPSVPLHSWPWPEEPWARLNLDFLGPFNNKYYLIIVDAHSKWIEVEKLSCTSATAVIACLRRLFARFGLAKRVVTDNGPPFSSVEFRSYLSKNGIKHLPVAPYHPSSNGAAENAVKTIKRVLKKALIEKEDDNTALTKFLFMYRNSEHSTTGREPAVALFGRRLRGRLDLLRPNTSSIVREAQLASESYNAGTAGYREAAEGDSVLLQDFTQGKKKWTPGTIKNRSGPVTYRVTADDGRVHKRHIDQILISNNSRKSRYSLSKLNNDFETKSVNDSVGEARESPGMTGEKELNSSYDTGPESPQPVTPSHSPAPNVLQPSRQRRKAALTCIQRIKDQNY
Summary
Uniprot
A0A1B6E6W0
A0A1B6CK52
A0A131XQT5
A0A3S2PB62
A0A3P8Q506
A0A1A7YSF0
+ More
A0A3P8QD94 A0A3P8NHI5 A0A3Q1EQC9 A2TGR4 A2TGR5 A0A3P8VSE7 A0A267EVG4 A0A3P8NEQ7 A0A2S2PU23 A0A2I4C6V0 A0A1S3J391 J9L709 A0A1A7WUZ7 A0A267F699 A0A1Y1KIG3 A0A1Y1LR51 A0A3P8RGT3 A0A267FRX0 A0A2R2MK13 A0A1B6H651 A0A1X7TH22 A0A267F772 A0A3B3H2N7 A0A267GNB6 A0A1X7TIE8 A0A1Y1N2I4 A0A3B3B4L9 A0A1Y1KCE7 A0A1S3KHX3 Q5KTM2 A0A147ADQ5 A0A1Y1NFA4 A0A3S0ZSV8 A0A3P8NEP7 A0A0V1M0R9 A0A2S2P240 A0A0V0RTT5 A0A146M835 A0A3P8NV61 A0A1Y1NIP2 A0A267GR92 A0A085LTS7 A0A267EXJ4 X1WXW3 A0A2R5LIR9 A0A085M8Q0 A0A146NT21 A0A146XMZ2 J9L9M6 A0A0V0TT39 A0A0V1BVI1 A0A1Y1LWB5 K7JW34 A0A1Y1ML73 A0A2S2PLE8 T2MJH3 A0A0V1LU74 A0A1Y1MAK2 A0A0V0VH28 A0A0V1GSV8 A0A0V1MIE6 A0A1S3SSU6 A0A3R7DF32 A0A0V0USW3 A0A1A8QZH2 A0A0V0Z754 A0A1Y1M6T5 A0A1S3HLN2 A0A1X7VU64 A0A085LME0 A0A0V1HJS5 A0A0V0ZP51 A0A1Y1M058 A0A0V0RYP2 A0A0V1KNI7 A0A0V1M3Q7 A0A085MHF4 A0A2H8TQH2 A0A085LS29 A0A0A9X8H3 A0A147BQ83 A0A147BJS0 A0A2G8LB01 A0A1S3JKW9 A0A1Y1K5V5 A0A1Y1K0Y3 A0A267EMN7 J9JQE4 A0A267DYU7 A0A085LXT4 A0A131Z1D5 A0A0V0T3M4 A0A1W7R696
A0A3P8QD94 A0A3P8NHI5 A0A3Q1EQC9 A2TGR4 A2TGR5 A0A3P8VSE7 A0A267EVG4 A0A3P8NEQ7 A0A2S2PU23 A0A2I4C6V0 A0A1S3J391 J9L709 A0A1A7WUZ7 A0A267F699 A0A1Y1KIG3 A0A1Y1LR51 A0A3P8RGT3 A0A267FRX0 A0A2R2MK13 A0A1B6H651 A0A1X7TH22 A0A267F772 A0A3B3H2N7 A0A267GNB6 A0A1X7TIE8 A0A1Y1N2I4 A0A3B3B4L9 A0A1Y1KCE7 A0A1S3KHX3 Q5KTM2 A0A147ADQ5 A0A1Y1NFA4 A0A3S0ZSV8 A0A3P8NEP7 A0A0V1M0R9 A0A2S2P240 A0A0V0RTT5 A0A146M835 A0A3P8NV61 A0A1Y1NIP2 A0A267GR92 A0A085LTS7 A0A267EXJ4 X1WXW3 A0A2R5LIR9 A0A085M8Q0 A0A146NT21 A0A146XMZ2 J9L9M6 A0A0V0TT39 A0A0V1BVI1 A0A1Y1LWB5 K7JW34 A0A1Y1ML73 A0A2S2PLE8 T2MJH3 A0A0V1LU74 A0A1Y1MAK2 A0A0V0VH28 A0A0V1GSV8 A0A0V1MIE6 A0A1S3SSU6 A0A3R7DF32 A0A0V0USW3 A0A1A8QZH2 A0A0V0Z754 A0A1Y1M6T5 A0A1S3HLN2 A0A1X7VU64 A0A085LME0 A0A0V1HJS5 A0A0V0ZP51 A0A1Y1M058 A0A0V0RYP2 A0A0V1KNI7 A0A0V1M3Q7 A0A085MHF4 A0A2H8TQH2 A0A085LS29 A0A0A9X8H3 A0A147BQ83 A0A147BJS0 A0A2G8LB01 A0A1S3JKW9 A0A1Y1K5V5 A0A1Y1K0Y3 A0A267EMN7 J9JQE4 A0A267DYU7 A0A085LXT4 A0A131Z1D5 A0A0V0T3M4 A0A1W7R696
Pubmed
EMBL
GEDC01007675
GEDC01003661
JAS29623.1
JAS33637.1
GEDC01023506
GEDC01023205
+ More
JAS13792.1 JAS14093.1 GEFM01006397 JAP69399.1 RSAL01000130 RVE46453.1 HADX01010638 SBP32870.1 EF199621 ABM90392.1 EF199622 ABM90393.1 NIVC01001652 PAA65446.1 GGMR01020219 MBY32838.1 ABLF02016687 ABLF02016690 HADW01008367 SBP09767.1 NIVC01001377 PAA68677.1 GEZM01085308 JAV60010.1 GEZM01050895 JAV75318.1 NIVC01000860 PAA75827.1 GECU01037588 JAS70118.1 NIVC01001308 PAA69606.1 NIVC01000264 PAA86792.1 GEZM01014356 JAV92133.1 GEZM01092768 GEZM01092767 JAV56467.1 AB126055 BAD86655.1 GCES01009650 JAR76673.1 GEZM01009753 JAV94337.1 RQTK01000185 RUS85038.1 JYDO01000377 KRZ65389.1 GGMR01010825 MBY23444.1 JYDL01000080 KRX17894.1 GDHC01002706 JAQ15923.1 GEZM01001427 JAV97782.1 NIVC01000183 PAA88548.1 KL363295 KFD48373.1 NIVC01001575 PAA66240.1 ABLF02006499 ABLF02041879 GGLE01005288 MBY09414.1 KL363215 KFD53596.1 GCES01151725 JAQ34597.1 GCES01042893 JAR43430.1 ABLF02065955 JYDJ01000151 KRX42156.1 JYDH01000010 KRY40903.1 GEZM01050896 JAV75317.1 AAZX01023237 GEZM01030100 JAV85808.1 GGMR01017626 MBY30245.1 HAAD01006009 CDG72241.1 JYDW01000004 KRZ62956.1 GEZM01036245 JAV82862.1 JYDN01000036 KRX62850.1 JYDP01000327 KRZ01162.1 JYDO01000093 KRZ71620.1 NIRI01001153 RJW67559.1 JYDN01000181 KRX54093.1 HAEH01013823 SBR98409.1 JYDQ01000346 KRY08270.1 GEZM01042258 JAV80005.1 KL363391 KFD46136.1 JYDP01000057 KRZ10689.1 JYDQ01000124 KRY14116.1 GEZM01043158 JAV79264.1 JYDL01000058 KRX19511.1 JYDW01000348 KRZ48953.1 JYDO01000236 KRZ66548.1 KL363192 KFD56650.1 GFXV01004590 MBW16395.1 KL363314 KFD47775.1 GBHO01027669 GBHO01027668 JAG15935.1 JAG15936.1 GEGO01002490 JAR92914.1 GEGO01004367 JAR91037.1 MRZV01000143 PIK57404.1 GEZM01096183 JAV55075.1 GEZM01096184 JAV55073.1 NIVC01001899 PAA62793.1 ABLF02011319 ABLF02014651 ABLF02041920 NIVC01003024 PAA53787.1 KL363264 KFD49780.1 GEDV01003323 JAP85234.1 JYDJ01000764 KRX33566.1 GEHC01000992 JAV46653.1
JAS13792.1 JAS14093.1 GEFM01006397 JAP69399.1 RSAL01000130 RVE46453.1 HADX01010638 SBP32870.1 EF199621 ABM90392.1 EF199622 ABM90393.1 NIVC01001652 PAA65446.1 GGMR01020219 MBY32838.1 ABLF02016687 ABLF02016690 HADW01008367 SBP09767.1 NIVC01001377 PAA68677.1 GEZM01085308 JAV60010.1 GEZM01050895 JAV75318.1 NIVC01000860 PAA75827.1 GECU01037588 JAS70118.1 NIVC01001308 PAA69606.1 NIVC01000264 PAA86792.1 GEZM01014356 JAV92133.1 GEZM01092768 GEZM01092767 JAV56467.1 AB126055 BAD86655.1 GCES01009650 JAR76673.1 GEZM01009753 JAV94337.1 RQTK01000185 RUS85038.1 JYDO01000377 KRZ65389.1 GGMR01010825 MBY23444.1 JYDL01000080 KRX17894.1 GDHC01002706 JAQ15923.1 GEZM01001427 JAV97782.1 NIVC01000183 PAA88548.1 KL363295 KFD48373.1 NIVC01001575 PAA66240.1 ABLF02006499 ABLF02041879 GGLE01005288 MBY09414.1 KL363215 KFD53596.1 GCES01151725 JAQ34597.1 GCES01042893 JAR43430.1 ABLF02065955 JYDJ01000151 KRX42156.1 JYDH01000010 KRY40903.1 GEZM01050896 JAV75317.1 AAZX01023237 GEZM01030100 JAV85808.1 GGMR01017626 MBY30245.1 HAAD01006009 CDG72241.1 JYDW01000004 KRZ62956.1 GEZM01036245 JAV82862.1 JYDN01000036 KRX62850.1 JYDP01000327 KRZ01162.1 JYDO01000093 KRZ71620.1 NIRI01001153 RJW67559.1 JYDN01000181 KRX54093.1 HAEH01013823 SBR98409.1 JYDQ01000346 KRY08270.1 GEZM01042258 JAV80005.1 KL363391 KFD46136.1 JYDP01000057 KRZ10689.1 JYDQ01000124 KRY14116.1 GEZM01043158 JAV79264.1 JYDL01000058 KRX19511.1 JYDW01000348 KRZ48953.1 JYDO01000236 KRZ66548.1 KL363192 KFD56650.1 GFXV01004590 MBW16395.1 KL363314 KFD47775.1 GBHO01027669 GBHO01027668 JAG15935.1 JAG15936.1 GEGO01002490 JAR92914.1 GEGO01004367 JAR91037.1 MRZV01000143 PIK57404.1 GEZM01096183 JAV55075.1 GEZM01096184 JAV55073.1 NIVC01001899 PAA62793.1 ABLF02011319 ABLF02014651 ABLF02041920 NIVC01003024 PAA53787.1 KL363264 KFD49780.1 GEDV01003323 JAP85234.1 JYDJ01000764 KRX33566.1 GEHC01000992 JAV46653.1
Proteomes
PRIDE
Pfam
Interpro
IPR001878
Znf_CCHC
+ More
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR041373 RT_RNaseH
IPR034128 K02A2.6-like
IPR001969 Aspartic_peptidase_AS
IPR018061 Retropepsins
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR005162 Retrotrans_gag_dom
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR041373 RT_RNaseH
IPR034128 K02A2.6-like
IPR001969 Aspartic_peptidase_AS
IPR018061 Retropepsins
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR005162 Retrotrans_gag_dom
Gene 3D
ProteinModelPortal
A0A1B6E6W0
A0A1B6CK52
A0A131XQT5
A0A3S2PB62
A0A3P8Q506
A0A1A7YSF0
+ More
A0A3P8QD94 A0A3P8NHI5 A0A3Q1EQC9 A2TGR4 A2TGR5 A0A3P8VSE7 A0A267EVG4 A0A3P8NEQ7 A0A2S2PU23 A0A2I4C6V0 A0A1S3J391 J9L709 A0A1A7WUZ7 A0A267F699 A0A1Y1KIG3 A0A1Y1LR51 A0A3P8RGT3 A0A267FRX0 A0A2R2MK13 A0A1B6H651 A0A1X7TH22 A0A267F772 A0A3B3H2N7 A0A267GNB6 A0A1X7TIE8 A0A1Y1N2I4 A0A3B3B4L9 A0A1Y1KCE7 A0A1S3KHX3 Q5KTM2 A0A147ADQ5 A0A1Y1NFA4 A0A3S0ZSV8 A0A3P8NEP7 A0A0V1M0R9 A0A2S2P240 A0A0V0RTT5 A0A146M835 A0A3P8NV61 A0A1Y1NIP2 A0A267GR92 A0A085LTS7 A0A267EXJ4 X1WXW3 A0A2R5LIR9 A0A085M8Q0 A0A146NT21 A0A146XMZ2 J9L9M6 A0A0V0TT39 A0A0V1BVI1 A0A1Y1LWB5 K7JW34 A0A1Y1ML73 A0A2S2PLE8 T2MJH3 A0A0V1LU74 A0A1Y1MAK2 A0A0V0VH28 A0A0V1GSV8 A0A0V1MIE6 A0A1S3SSU6 A0A3R7DF32 A0A0V0USW3 A0A1A8QZH2 A0A0V0Z754 A0A1Y1M6T5 A0A1S3HLN2 A0A1X7VU64 A0A085LME0 A0A0V1HJS5 A0A0V0ZP51 A0A1Y1M058 A0A0V0RYP2 A0A0V1KNI7 A0A0V1M3Q7 A0A085MHF4 A0A2H8TQH2 A0A085LS29 A0A0A9X8H3 A0A147BQ83 A0A147BJS0 A0A2G8LB01 A0A1S3JKW9 A0A1Y1K5V5 A0A1Y1K0Y3 A0A267EMN7 J9JQE4 A0A267DYU7 A0A085LXT4 A0A131Z1D5 A0A0V0T3M4 A0A1W7R696
A0A3P8QD94 A0A3P8NHI5 A0A3Q1EQC9 A2TGR4 A2TGR5 A0A3P8VSE7 A0A267EVG4 A0A3P8NEQ7 A0A2S2PU23 A0A2I4C6V0 A0A1S3J391 J9L709 A0A1A7WUZ7 A0A267F699 A0A1Y1KIG3 A0A1Y1LR51 A0A3P8RGT3 A0A267FRX0 A0A2R2MK13 A0A1B6H651 A0A1X7TH22 A0A267F772 A0A3B3H2N7 A0A267GNB6 A0A1X7TIE8 A0A1Y1N2I4 A0A3B3B4L9 A0A1Y1KCE7 A0A1S3KHX3 Q5KTM2 A0A147ADQ5 A0A1Y1NFA4 A0A3S0ZSV8 A0A3P8NEP7 A0A0V1M0R9 A0A2S2P240 A0A0V0RTT5 A0A146M835 A0A3P8NV61 A0A1Y1NIP2 A0A267GR92 A0A085LTS7 A0A267EXJ4 X1WXW3 A0A2R5LIR9 A0A085M8Q0 A0A146NT21 A0A146XMZ2 J9L9M6 A0A0V0TT39 A0A0V1BVI1 A0A1Y1LWB5 K7JW34 A0A1Y1ML73 A0A2S2PLE8 T2MJH3 A0A0V1LU74 A0A1Y1MAK2 A0A0V0VH28 A0A0V1GSV8 A0A0V1MIE6 A0A1S3SSU6 A0A3R7DF32 A0A0V0USW3 A0A1A8QZH2 A0A0V0Z754 A0A1Y1M6T5 A0A1S3HLN2 A0A1X7VU64 A0A085LME0 A0A0V1HJS5 A0A0V0ZP51 A0A1Y1M058 A0A0V0RYP2 A0A0V1KNI7 A0A0V1M3Q7 A0A085MHF4 A0A2H8TQH2 A0A085LS29 A0A0A9X8H3 A0A147BQ83 A0A147BJS0 A0A2G8LB01 A0A1S3JKW9 A0A1Y1K5V5 A0A1Y1K0Y3 A0A267EMN7 J9JQE4 A0A267DYU7 A0A085LXT4 A0A131Z1D5 A0A0V0T3M4 A0A1W7R696
PDB
4OL8
E-value=5.3887e-53,
Score=530
Ontologies
KEGG
GO
Topology
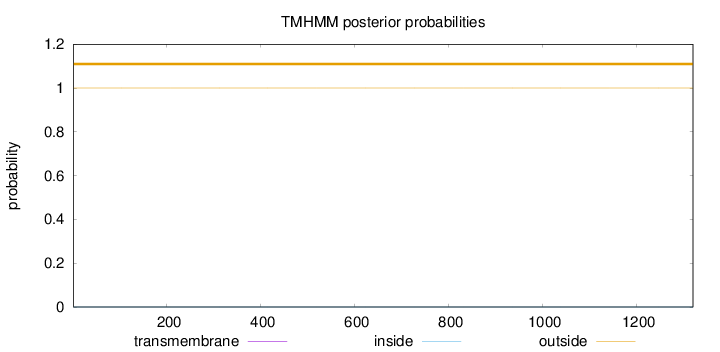
Length:
1320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000800000000000001
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00002
outside
1 - 1320
Population Genetic Test Statistics
Pi
15.959026
Theta
18.808697
Tajima's D
-0.949986
CLR
0
CSRT
0.147742612869357
Interpretation
Uncertain
