Gene
KWMTBOMO16533
Pre Gene Modal
BGIBMGA005562
Annotation
hypothetical_protein_KGM_04574_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.895
Sequence
CDS
ATGGCGGTGTTCGTGTTCTGGTGCTACGCGTGCAAGTTCGTGCCGGCCGCCGTCTCCATCCTCGTGCTGTACGGGCTGACGCTGCGCAGCTTCTGCGAGCAGATCGCGGCCGCCACCAACGCGACGTCGTGCTGGATCGTGTACCCGGTGAACGTGGGCGCGTTCCCGCCCAGCCACGACCTCACCGCGAGGAGCTGGCACGGCTGGGGCGACGACTTCTACGACGGATTCTATTTGACCCAACATATCGTGCTCGCTTTGATAACTTTGCATTTAATTGTGATCTCATTATCATTCGTCCATCGCTACTCGTCGATATGGCAACGTGGCTTCTGGACCAATCGGGCGTGGACCATCACTGTTGTCGTTCTGCTAACCGCTCCGCACTCCCGCAGACTGGTAGTCCAGTGCGCGTTCAGCGGCGTGATGCTGAGCGGCACGTACGGGTCGTGCGCGCGCTGGTCGGTGTGCGCGCTGCGGTACCGCGTGCCGTGGCCGCTGCTGCTCGCGTACGCCGCCTCGCTGCTGCTCGTGCTCGCGCTCAACGAGCTCATCAAGTGGCAGGAGATCAAGGCAGACAAACGGAATCAACGCCGCGCCCGTCTCGACTTCGGAACCAAACTGGGAATGAATTCTCCATTCTAA
Protein
MAVFVFWCYACKFVPAAVSILVLYGLTLRSFCEQIAAATNATSCWIVYPVNVGAFPPSHDLTARSWHGWGDDFYDGFYLTQHIVLALITLHLIVISLSFVHRYSSIWQRGFWTNRAWTITVVVLLTAPHSRRLVVQCAFSGVMLSGTYGSCARWSVCALRYRVPWPLLLAYAASLLLVLALNELIKWQEIKADKRNQRRARLDFGTKLGMNSPF
Summary
Uniprot
A0A2A4JEV8
A0A3S2L8H5
A0A194QB23
A0A194QSL0
A0A212EPB0
A0A2J7PXI5
+ More
A0A2J7PXH2 A0A2J7PXH6 A0A067QXM3 A0A2J7PXG9 A0A1B6DN27 A0A1B6BXT8 A0A1B6EF44 A0A224XDD0 A0A023F1Q2 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DTF3 A0A1B6DHS1 A0A310SMW2 A0A087ZXX4 A0A2A3EAG4 A0A2R7VPD4 A0A232ER02 A0A0N0U4H9 T1HGD1 A0A0C9RMK5 A0A0L7QQV6 A0A0J7K7I4 E2B474 A0A0C9QMF3 A0A0C9PL89 A0A151JXP4 A0A195CWZ6 E1ZV00 A0A151XIG3 A0A026WG49 A0A3L8DJS8 F4WHR9 R7TYH0 E0VQ73 A0A195B5D9 A0A158NM24 A0A1L8D957 T1IVZ1 A0A1Y1JWC3 A0A1Y1JTV9 A0A1Y1JTB8 A0A1Y1JYJ7 A0A0A9Y593 A0A2P8XCV9 A0A1Y1JXQ8 A0A1Y1JXH7 A0A0A9XA88 A0A146LYF4 A0A146M4X0 A0A146LUX9 A0A0P5SYX3 A0A1B6JBX6 A0A1B6HLI4 A0A2S2P6G4 A0A2H8TVQ2 A0A0P5X8W6 G1PRY8 A0A0P5Y0B1 A0A0P5XJJ0 L5LY92 A0A0P5NIP6 A0A0N8CZF9 A0A0P5WK49 A0A0N8B0W7 A0A0P5Q8T5 A0A0P6H8G9 A0A0N8CES9 A0A0P6HDN5 A0A0P5HU79 A0A3Q7SGT7 E9HGS6 A0A3Q7S7V7 A0A3Q7T5T7 A0A3Q7SGU2 A0A3Q7TK60 A0A3Q7S7V3 A0A3Q7SJG2 A0A3Q7T5T3 A0A3Q7T5U0 A0A3Q7TK66 A0A3Q7SJF6 A0A3Q7S7V0 A0A3Q7SGT1 J9NTK0 F1PD09 S7QCU6 K9IP41
A0A2J7PXH2 A0A2J7PXH6 A0A067QXM3 A0A2J7PXG9 A0A1B6DN27 A0A1B6BXT8 A0A1B6EF44 A0A224XDD0 A0A023F1Q2 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DTF3 A0A1B6DHS1 A0A310SMW2 A0A087ZXX4 A0A2A3EAG4 A0A2R7VPD4 A0A232ER02 A0A0N0U4H9 T1HGD1 A0A0C9RMK5 A0A0L7QQV6 A0A0J7K7I4 E2B474 A0A0C9QMF3 A0A0C9PL89 A0A151JXP4 A0A195CWZ6 E1ZV00 A0A151XIG3 A0A026WG49 A0A3L8DJS8 F4WHR9 R7TYH0 E0VQ73 A0A195B5D9 A0A158NM24 A0A1L8D957 T1IVZ1 A0A1Y1JWC3 A0A1Y1JTV9 A0A1Y1JTB8 A0A1Y1JYJ7 A0A0A9Y593 A0A2P8XCV9 A0A1Y1JXQ8 A0A1Y1JXH7 A0A0A9XA88 A0A146LYF4 A0A146M4X0 A0A146LUX9 A0A0P5SYX3 A0A1B6JBX6 A0A1B6HLI4 A0A2S2P6G4 A0A2H8TVQ2 A0A0P5X8W6 G1PRY8 A0A0P5Y0B1 A0A0P5XJJ0 L5LY92 A0A0P5NIP6 A0A0N8CZF9 A0A0P5WK49 A0A0N8B0W7 A0A0P5Q8T5 A0A0P6H8G9 A0A0N8CES9 A0A0P6HDN5 A0A0P5HU79 A0A3Q7SGT7 E9HGS6 A0A3Q7S7V7 A0A3Q7T5T7 A0A3Q7SGU2 A0A3Q7TK60 A0A3Q7S7V3 A0A3Q7SJG2 A0A3Q7T5T3 A0A3Q7T5U0 A0A3Q7TK66 A0A3Q7SJF6 A0A3Q7S7V0 A0A3Q7SGT1 J9NTK0 F1PD09 S7QCU6 K9IP41
Pubmed
EMBL
NWSH01001832
PCG69963.1
RSAL01000090
RVE48102.1
KQ459232
KPJ02629.1
+ More
KQ461155 KPJ08304.1 AGBW02013517 OWR43291.1 NEVH01020858 PNF21040.1 PNF21041.1 PNF21037.1 KK853134 KDR10862.1 PNF21038.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01000814 JAS36484.1 GFTR01008638 JAW07788.1 GBBI01003783 JAC14929.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01026348 JAS10950.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 KQ759847 OAD62585.1 KZ288308 PBC28715.1 KK854009 PTY09219.1 NNAY01002685 OXU20778.1 KQ435834 KOX71551.1 ACPB03015962 ACPB03015963 GBYB01009580 JAG79347.1 KQ414784 KOC61013.1 LBMM01012452 KMQ86214.1 GL445515 EFN89491.1 GBYB01001767 JAG71534.1 GBYB01001768 JAG71535.1 KQ981561 KYN39827.1 KQ977171 KYN05181.1 GL434335 EFN74998.1 KQ982109 KYQ60000.1 KK107238 EZA54621.1 QOIP01000007 RLU20670.1 GL888167 EGI66276.1 AMQN01011293 KB308811 ELT96471.1 DS235392 EEB15529.1 KQ976604 KYM79404.1 ADTU01020204 GFDF01011093 JAV02991.1 JH431601 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101579 GEZM01101575 GEZM01101571 GEZM01101568 GEZM01101562 GEZM01101560 GEZM01101559 GEZM01101558 GEZM01101555 GEZM01101554 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101542 GEZM01101541 GEZM01101540 JAV52532.1 GEZM01101578 JAV52495.1 GBHO01045648 GBHO01045647 GBHO01045646 GBHO01021173 GBHO01016799 GBHO01016798 JAF97955.1 JAF97956.1 JAF97957.1 JAG22431.1 JAG26805.1 JAG26806.1 PYGN01002942 PSN29814.1 GEZM01101544 JAV52590.1 GEZM01101577 GEZM01101572 GEZM01101563 GEZM01101561 GEZM01101552 GEZM01101543 JAV52510.1 GBHO01045650 GBHO01045645 GBHO01026686 GBHO01026683 GBHO01026682 GBHO01026681 GBHO01026680 GBHO01026678 JAF97953.1 JAF97958.1 JAG16918.1 JAG16921.1 JAG16922.1 JAG16923.1 JAG16924.1 JAG16926.1 GDHC01006927 JAQ11702.1 GDHC01003801 JAQ14828.1 GDHC01007108 JAQ11521.1 GDIP01133416 JAL70298.1 GECU01011041 JAS96665.1 GECU01032144 JAS75562.1 GGMR01012391 MBY25010.1 GFXV01006195 MBW18000.1 GDIP01087734 JAM15981.1 AAPE02047503 GDIP01064682 JAM39033.1 GDIP01071432 JAM32283.1 KB106579 ELK30970.1 GDIQ01144655 JAL07071.1 GDIP01083775 JAM19940.1 GDIP01085322 JAM18393.1 GDIQ01223723 JAK28002.1 GDIQ01117910 JAL33816.1 GDIQ01044793 JAN49944.1 GDIP01138815 JAL64899.1 GDIQ01020965 JAN73772.1 GDIQ01224974 JAK26751.1 GL732643 EFX69064.1 AAEX03006298 KE164590 EPQ18977.1 GABZ01003675 JAA49850.1
KQ461155 KPJ08304.1 AGBW02013517 OWR43291.1 NEVH01020858 PNF21040.1 PNF21041.1 PNF21037.1 KK853134 KDR10862.1 PNF21038.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01000814 JAS36484.1 GFTR01008638 JAW07788.1 GBBI01003783 JAC14929.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01026348 JAS10950.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 KQ759847 OAD62585.1 KZ288308 PBC28715.1 KK854009 PTY09219.1 NNAY01002685 OXU20778.1 KQ435834 KOX71551.1 ACPB03015962 ACPB03015963 GBYB01009580 JAG79347.1 KQ414784 KOC61013.1 LBMM01012452 KMQ86214.1 GL445515 EFN89491.1 GBYB01001767 JAG71534.1 GBYB01001768 JAG71535.1 KQ981561 KYN39827.1 KQ977171 KYN05181.1 GL434335 EFN74998.1 KQ982109 KYQ60000.1 KK107238 EZA54621.1 QOIP01000007 RLU20670.1 GL888167 EGI66276.1 AMQN01011293 KB308811 ELT96471.1 DS235392 EEB15529.1 KQ976604 KYM79404.1 ADTU01020204 GFDF01011093 JAV02991.1 JH431601 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101579 GEZM01101575 GEZM01101571 GEZM01101568 GEZM01101562 GEZM01101560 GEZM01101559 GEZM01101558 GEZM01101555 GEZM01101554 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101542 GEZM01101541 GEZM01101540 JAV52532.1 GEZM01101578 JAV52495.1 GBHO01045648 GBHO01045647 GBHO01045646 GBHO01021173 GBHO01016799 GBHO01016798 JAF97955.1 JAF97956.1 JAF97957.1 JAG22431.1 JAG26805.1 JAG26806.1 PYGN01002942 PSN29814.1 GEZM01101544 JAV52590.1 GEZM01101577 GEZM01101572 GEZM01101563 GEZM01101561 GEZM01101552 GEZM01101543 JAV52510.1 GBHO01045650 GBHO01045645 GBHO01026686 GBHO01026683 GBHO01026682 GBHO01026681 GBHO01026680 GBHO01026678 JAF97953.1 JAF97958.1 JAG16918.1 JAG16921.1 JAG16922.1 JAG16923.1 JAG16924.1 JAG16926.1 GDHC01006927 JAQ11702.1 GDHC01003801 JAQ14828.1 GDHC01007108 JAQ11521.1 GDIP01133416 JAL70298.1 GECU01011041 JAS96665.1 GECU01032144 JAS75562.1 GGMR01012391 MBY25010.1 GFXV01006195 MBW18000.1 GDIP01087734 JAM15981.1 AAPE02047503 GDIP01064682 JAM39033.1 GDIP01071432 JAM32283.1 KB106579 ELK30970.1 GDIQ01144655 JAL07071.1 GDIP01083775 JAM19940.1 GDIP01085322 JAM18393.1 GDIQ01223723 JAK28002.1 GDIQ01117910 JAL33816.1 GDIQ01044793 JAN49944.1 GDIP01138815 JAL64899.1 GDIQ01020965 JAN73772.1 GDIQ01224974 JAK26751.1 GL732643 EFX69064.1 AAEX03006298 KE164590 EPQ18977.1 GABZ01003675 JAA49850.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000027135 UP000005203 UP000242457 UP000215335 UP000053105 UP000015103 UP000053825 UP000036403 UP000008237 UP000078541 UP000078542 UP000000311 UP000075809 UP000053097 UP000279307 UP000007755 UP000014760 UP000009046 UP000078540 UP000005205 UP000245037 UP000001074 UP000286640 UP000000305 UP000002254
UP000027135 UP000005203 UP000242457 UP000215335 UP000053105 UP000015103 UP000053825 UP000036403 UP000008237 UP000078541 UP000078542 UP000000311 UP000075809 UP000053097 UP000279307 UP000007755 UP000014760 UP000009046 UP000078540 UP000005205 UP000245037 UP000001074 UP000286640 UP000000305 UP000002254
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JEV8
A0A3S2L8H5
A0A194QB23
A0A194QSL0
A0A212EPB0
A0A2J7PXI5
+ More
A0A2J7PXH2 A0A2J7PXH6 A0A067QXM3 A0A2J7PXG9 A0A1B6DN27 A0A1B6BXT8 A0A1B6EF44 A0A224XDD0 A0A023F1Q2 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DTF3 A0A1B6DHS1 A0A310SMW2 A0A087ZXX4 A0A2A3EAG4 A0A2R7VPD4 A0A232ER02 A0A0N0U4H9 T1HGD1 A0A0C9RMK5 A0A0L7QQV6 A0A0J7K7I4 E2B474 A0A0C9QMF3 A0A0C9PL89 A0A151JXP4 A0A195CWZ6 E1ZV00 A0A151XIG3 A0A026WG49 A0A3L8DJS8 F4WHR9 R7TYH0 E0VQ73 A0A195B5D9 A0A158NM24 A0A1L8D957 T1IVZ1 A0A1Y1JWC3 A0A1Y1JTV9 A0A1Y1JTB8 A0A1Y1JYJ7 A0A0A9Y593 A0A2P8XCV9 A0A1Y1JXQ8 A0A1Y1JXH7 A0A0A9XA88 A0A146LYF4 A0A146M4X0 A0A146LUX9 A0A0P5SYX3 A0A1B6JBX6 A0A1B6HLI4 A0A2S2P6G4 A0A2H8TVQ2 A0A0P5X8W6 G1PRY8 A0A0P5Y0B1 A0A0P5XJJ0 L5LY92 A0A0P5NIP6 A0A0N8CZF9 A0A0P5WK49 A0A0N8B0W7 A0A0P5Q8T5 A0A0P6H8G9 A0A0N8CES9 A0A0P6HDN5 A0A0P5HU79 A0A3Q7SGT7 E9HGS6 A0A3Q7S7V7 A0A3Q7T5T7 A0A3Q7SGU2 A0A3Q7TK60 A0A3Q7S7V3 A0A3Q7SJG2 A0A3Q7T5T3 A0A3Q7T5U0 A0A3Q7TK66 A0A3Q7SJF6 A0A3Q7S7V0 A0A3Q7SGT1 J9NTK0 F1PD09 S7QCU6 K9IP41
A0A2J7PXH2 A0A2J7PXH6 A0A067QXM3 A0A2J7PXG9 A0A1B6DN27 A0A1B6BXT8 A0A1B6EF44 A0A224XDD0 A0A023F1Q2 A0A1B6BYR1 A0A1B6CCC8 A0A1B6DTF3 A0A1B6DHS1 A0A310SMW2 A0A087ZXX4 A0A2A3EAG4 A0A2R7VPD4 A0A232ER02 A0A0N0U4H9 T1HGD1 A0A0C9RMK5 A0A0L7QQV6 A0A0J7K7I4 E2B474 A0A0C9QMF3 A0A0C9PL89 A0A151JXP4 A0A195CWZ6 E1ZV00 A0A151XIG3 A0A026WG49 A0A3L8DJS8 F4WHR9 R7TYH0 E0VQ73 A0A195B5D9 A0A158NM24 A0A1L8D957 T1IVZ1 A0A1Y1JWC3 A0A1Y1JTV9 A0A1Y1JTB8 A0A1Y1JYJ7 A0A0A9Y593 A0A2P8XCV9 A0A1Y1JXQ8 A0A1Y1JXH7 A0A0A9XA88 A0A146LYF4 A0A146M4X0 A0A146LUX9 A0A0P5SYX3 A0A1B6JBX6 A0A1B6HLI4 A0A2S2P6G4 A0A2H8TVQ2 A0A0P5X8W6 G1PRY8 A0A0P5Y0B1 A0A0P5XJJ0 L5LY92 A0A0P5NIP6 A0A0N8CZF9 A0A0P5WK49 A0A0N8B0W7 A0A0P5Q8T5 A0A0P6H8G9 A0A0N8CES9 A0A0P6HDN5 A0A0P5HU79 A0A3Q7SGT7 E9HGS6 A0A3Q7S7V7 A0A3Q7T5T7 A0A3Q7SGU2 A0A3Q7TK60 A0A3Q7S7V3 A0A3Q7SJG2 A0A3Q7T5T3 A0A3Q7T5U0 A0A3Q7TK66 A0A3Q7SJF6 A0A3Q7S7V0 A0A3Q7SGT1 J9NTK0 F1PD09 S7QCU6 K9IP41
Ontologies
PANTHER
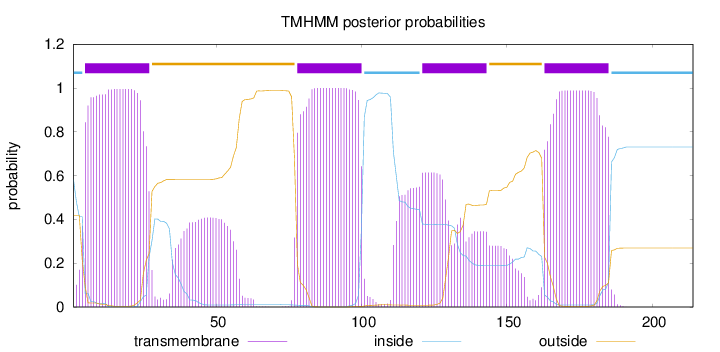
Topology
Length:
214
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
93.62219
Exp number, first 60 AAs:
31.01757
Total prob of N-in:
0.58165
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 77
TMhelix
78 - 100
inside
101 - 120
TMhelix
121 - 143
outside
144 - 162
TMhelix
163 - 185
inside
186 - 214
Population Genetic Test Statistics
Pi
30.524814
Theta
31.249941
Tajima's D
-0.797796
CLR
0
CSRT
0.175041247937603
Interpretation
Uncertain
