Gene
KWMTBOMO16529
Pre Gene Modal
BGIBMGA005639
Annotation
PREDICTED:_DNA_fragmentation_factor_subunit_beta_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.972
Sequence
CDS
ATGAAGAAAGGTTATAAAGTGACTGGTGTTAAACGAGAAAAGAAAATTGGCGTGGCAGCTGAAAGTTTACACGAGTTGATCGAGAAATCACACAAAAAATTAGAATTCAATGTCAGCTGCGGAGAATGTAATTTATACGTGGCTGAAGATGGGACGCTAGTTGACAATGATGAGTACCTAGCCACTCTGCCACCACAAACCGTCTTCATTTTACTAAAGGGTTCAGAAAAGATGATAACAGATTTTGATTATTATTACAATATGATCCTGTCAACTAAGAAAGAGTATATAGATACAGGAGCAGCAGCCAAGCAGTTTTTATCGACAAACATCAAAGAGAAGTTCAAAGTCTTTCAAAAGTACATTGCATCTGCTGACGATGCTAGAATAGAGCTCAGCGAGAGAACACAAGATCCAGCTTGGTTTGAAGGTCTTGAACCCTCTGAGAAAACAAAAGAGCAATCAATGTCTAAACGGGTCAAAGAACGAATGCGTGGCTACTATTATAAGACGAAATCAGCACTGCAGTCTTCCGAAACATATGTGCAATCCCGCAACGGACGCGGTAAGAAAATTATCGATCAGTTTTTGTTAGATCTGCGCAAACTACTTGAATCGAACAAGTATAACGAGGACTATTTCAATCGTAAAGCAGACAAAAGCGTCCGAATGTGCAATGTGACTGGTCTCTTTGAATGCGGTGGTTTATGGAACAATAACAAATGTGCCTATGAAGGTGACCACATTATCAACCCATACCGGAGCCGCGAGGAACGAATCATATTCCAGACATGGAATTTGGATCACAAAATTGAACTCTCCCGTTCAATAATACCTAAAATTCTGGAAGCCATAGAAGTTTTGTATAAGGGTGATTTTAAATGTACAGTGTGCGACAAATACTCTAAACCGGGTAGCATTGATACAGATAGGTATTATCTGCAAATCTTTACAAAAAACAACCTCAAACTGGTTCACATTGTATGTCATCATAAGGGCAGGCACAATGCTGCCTCTGACGTTTACACTGTCTGCAACAATTGTAGCAGACATACTATTGAATATATATGA
Protein
MKKGYKVTGVKREKKIGVAAESLHELIEKSHKKLEFNVSCGECNLYVAEDGTLVDNDEYLATLPPQTVFILLKGSEKMITDFDYYYNMILSTKKEYIDTGAAAKQFLSTNIKEKFKVFQKYIASADDARIELSERTQDPAWFEGLEPSEKTKEQSMSKRVKERMRGYYYKTKSALQSSETYVQSRNGRGKKIIDQFLLDLRKLLESNKYNEDYFNRKADKSVRMCNVTGLFECGGLWNNNKCAYEGDHIINPYRSREERIIFQTWNLDHKIELSRSIIPKILEAIEVLYKGDFKCTVCDKYSKPGSIDTDRYYLQIFTKNNLKLVHIVCHHKGRHNAASDVYTVCNNCSRHTIEYI
Summary
Uniprot
A0A2H1VMR1
A0A1E1WJU4
L7X1G4
D0AB91
A0A194QS01
A0A0L7LGR6
+ More
A0A212EP59 A0A067QYQ3 A0A146LJ53 A0A2J7PXZ6 A0A146LWA4 D6WW09 A0A2P8ZLG4 A0A1B6KTR6 A0A1B6EA14 A0A154PA95 W5JGP2 A0A182J889 A0A0M9A9K2 U4U402 N6SU96 A0A182M0H1 A0A182VTZ2 A0A182NBH5 A0A182R3W6 J9KG38 A0A023F0S3 A0A310SHW6 A0A2S2QP19 A0A224XFN9 A0A0T6AZL6 A0A1B0ETL7 A0A069DSR6 A0A0P4VIT5 A0A2R7WW04 A0A0L7RDP7 A0A026VZ81 A0A1Y1KI19 A0A1B6IS96 A0A1B6H5P9 A0A195CRW1 A0A151XAD2 A0A3M6UJD4 A0A195EDW5 A0A151I3U3 C3YR00 A0A2B4RSP0 A0A1A8UA31 A0A1A7ZUX4 A0A1A8CD96 A0A3B3HC86 A0A1S3JSC1 E0VAJ8 A0A3P9LXE9 A0A1A8F3T9 A0A3B5KTP5 A0A1A7XMY2 M3ZSJ8 A0A1A8IYQ6 A0A1A8PFK5 A0A3Q2PQP1 A0A3B3CCN6 A0A3P9JSC5 A0A3B4E6T0 A0A3B5RC92 A0A3B5L6R9 W5LKE6 A0A3S2MZ49 A0A3B4ZY20 A0A146QWA2 A0A3Q2CW62 A0A3Q2PP69 A0A315WBV3 A0A2D0QWW1 A0A3Q2ZNK3 I3JHL3 A0A3B3UWV6 A0A3B3YXE0 A0A087X8J9 A0A3P9P2A0 A0A3Q3MMN0 A0A3Q2QVN8 A0A3Q3M567 A0A147B0E9 A0A0S7EI22 A0A2I4C784 A0A146PQC1 Q9DGL7 Q6NWG2 A0A210QD94 A0A3P8Q9I1 A0A3P9D373 A0A3Q3MAF1 A0A0B7BH20 Q1L9C6 A0A3Q1HMM0 A0A3Q2XWN8
A0A212EP59 A0A067QYQ3 A0A146LJ53 A0A2J7PXZ6 A0A146LWA4 D6WW09 A0A2P8ZLG4 A0A1B6KTR6 A0A1B6EA14 A0A154PA95 W5JGP2 A0A182J889 A0A0M9A9K2 U4U402 N6SU96 A0A182M0H1 A0A182VTZ2 A0A182NBH5 A0A182R3W6 J9KG38 A0A023F0S3 A0A310SHW6 A0A2S2QP19 A0A224XFN9 A0A0T6AZL6 A0A1B0ETL7 A0A069DSR6 A0A0P4VIT5 A0A2R7WW04 A0A0L7RDP7 A0A026VZ81 A0A1Y1KI19 A0A1B6IS96 A0A1B6H5P9 A0A195CRW1 A0A151XAD2 A0A3M6UJD4 A0A195EDW5 A0A151I3U3 C3YR00 A0A2B4RSP0 A0A1A8UA31 A0A1A7ZUX4 A0A1A8CD96 A0A3B3HC86 A0A1S3JSC1 E0VAJ8 A0A3P9LXE9 A0A1A8F3T9 A0A3B5KTP5 A0A1A7XMY2 M3ZSJ8 A0A1A8IYQ6 A0A1A8PFK5 A0A3Q2PQP1 A0A3B3CCN6 A0A3P9JSC5 A0A3B4E6T0 A0A3B5RC92 A0A3B5L6R9 W5LKE6 A0A3S2MZ49 A0A3B4ZY20 A0A146QWA2 A0A3Q2CW62 A0A3Q2PP69 A0A315WBV3 A0A2D0QWW1 A0A3Q2ZNK3 I3JHL3 A0A3B3UWV6 A0A3B3YXE0 A0A087X8J9 A0A3P9P2A0 A0A3Q3MMN0 A0A3Q2QVN8 A0A3Q3M567 A0A147B0E9 A0A0S7EI22 A0A2I4C784 A0A146PQC1 Q9DGL7 Q6NWG2 A0A210QD94 A0A3P8Q9I1 A0A3P9D373 A0A3Q3MAF1 A0A0B7BH20 Q1L9C6 A0A3Q1HMM0 A0A3Q2XWN8
Pubmed
EMBL
ODYU01003152
SOQ41544.1
GDQN01003780
JAT87274.1
KC469893
AGC92696.1
+ More
FP102340 CBH09286.1 KQ461155 KPJ08298.1 JTDY01001151 KOB74728.1 AGBW02013517 OWR43285.1 KK852819 KDR15630.1 GDHC01011687 JAQ06942.1 NEVH01020852 PNF21196.1 GDHC01006685 JAQ11944.1 KQ971361 EFA08645.1 PYGN01000023 PSN57344.1 GEBQ01025132 JAT14845.1 GEDC01002535 JAS34763.1 KQ434856 KZC08761.1 ADMH02001555 ETN62080.1 KQ435720 KOX78649.1 KB631617 ERL84705.1 APGK01057068 KB741277 ENN71274.1 AXCM01005301 ABLF02033238 ABLF02033247 ABLF02033251 GBBI01003642 JAC15070.1 KQ768821 OAD53042.1 GGMS01010274 MBY79477.1 GFTR01005141 JAW11285.1 LJIG01016429 KRT80627.1 AJWK01011181 GBGD01002118 JAC86771.1 GDKW01001791 JAI54804.1 KK855487 PTY22785.1 KQ414614 KOC68920.1 KK107672 EZA48154.1 GEZM01083778 GEZM01083776 GEZM01083775 JAV61014.1 GECU01017939 JAS89767.1 GECU01037677 JAS70029.1 KQ977372 KYN03232.1 KQ982351 KYQ57238.1 RCHS01001413 RMX53745.1 KQ979039 KYN23398.1 KQ976475 KYM83920.1 GG666545 EEN57242.1 LSMT01000368 PFX19275.1 HAEJ01003516 SBS43973.1 HADY01007738 SBP46223.1 HADZ01013866 SBP77807.1 DS235008 EEB10404.1 HAEB01006242 SBQ52769.1 HADW01017958 SBP19358.1 HAED01015991 SBR02436.1 HAEH01006656 SBR79817.1 CM012443 RVE70103.1 GCES01125918 JAQ60404.1 NHOQ01000099 PWA32962.1 AERX01037009 AYCK01014051 GCES01002114 JAR84209.1 GBYX01476951 JAO04727.1 GCES01140179 JAQ46143.1 AF286179 BC092678 BC164850 AF426316 AAF99706.1 AAH92678.2 AAI64850.1 AAL40264.1 BC067602 AAH67602.1 NEDP02004099 OWF46705.1 HACG01044741 CEK91606.1 CR352301
FP102340 CBH09286.1 KQ461155 KPJ08298.1 JTDY01001151 KOB74728.1 AGBW02013517 OWR43285.1 KK852819 KDR15630.1 GDHC01011687 JAQ06942.1 NEVH01020852 PNF21196.1 GDHC01006685 JAQ11944.1 KQ971361 EFA08645.1 PYGN01000023 PSN57344.1 GEBQ01025132 JAT14845.1 GEDC01002535 JAS34763.1 KQ434856 KZC08761.1 ADMH02001555 ETN62080.1 KQ435720 KOX78649.1 KB631617 ERL84705.1 APGK01057068 KB741277 ENN71274.1 AXCM01005301 ABLF02033238 ABLF02033247 ABLF02033251 GBBI01003642 JAC15070.1 KQ768821 OAD53042.1 GGMS01010274 MBY79477.1 GFTR01005141 JAW11285.1 LJIG01016429 KRT80627.1 AJWK01011181 GBGD01002118 JAC86771.1 GDKW01001791 JAI54804.1 KK855487 PTY22785.1 KQ414614 KOC68920.1 KK107672 EZA48154.1 GEZM01083778 GEZM01083776 GEZM01083775 JAV61014.1 GECU01017939 JAS89767.1 GECU01037677 JAS70029.1 KQ977372 KYN03232.1 KQ982351 KYQ57238.1 RCHS01001413 RMX53745.1 KQ979039 KYN23398.1 KQ976475 KYM83920.1 GG666545 EEN57242.1 LSMT01000368 PFX19275.1 HAEJ01003516 SBS43973.1 HADY01007738 SBP46223.1 HADZ01013866 SBP77807.1 DS235008 EEB10404.1 HAEB01006242 SBQ52769.1 HADW01017958 SBP19358.1 HAED01015991 SBR02436.1 HAEH01006656 SBR79817.1 CM012443 RVE70103.1 GCES01125918 JAQ60404.1 NHOQ01000099 PWA32962.1 AERX01037009 AYCK01014051 GCES01002114 JAR84209.1 GBYX01476951 JAO04727.1 GCES01140179 JAQ46143.1 AF286179 BC092678 BC164850 AF426316 AAF99706.1 AAH92678.2 AAI64850.1 AAL40264.1 BC067602 AAH67602.1 NEDP02004099 OWF46705.1 HACG01044741 CEK91606.1 CR352301
Proteomes
UP000053240
UP000037510
UP000007151
UP000027135
UP000235965
UP000007266
+ More
UP000245037 UP000076502 UP000000673 UP000075880 UP000053105 UP000030742 UP000019118 UP000075883 UP000075920 UP000075884 UP000075900 UP000007819 UP000092461 UP000053825 UP000053097 UP000078542 UP000075809 UP000275408 UP000078492 UP000078540 UP000001554 UP000225706 UP000001038 UP000085678 UP000009046 UP000265180 UP000261380 UP000002852 UP000265000 UP000261560 UP000265200 UP000261440 UP000018467 UP000261400 UP000265020 UP000221080 UP000264800 UP000005207 UP000261500 UP000261480 UP000028760 UP000242638 UP000261640 UP000192220 UP000242188 UP000265100 UP000265160 UP000000437 UP000265040 UP000264820
UP000245037 UP000076502 UP000000673 UP000075880 UP000053105 UP000030742 UP000019118 UP000075883 UP000075920 UP000075884 UP000075900 UP000007819 UP000092461 UP000053825 UP000053097 UP000078542 UP000075809 UP000275408 UP000078492 UP000078540 UP000001554 UP000225706 UP000001038 UP000085678 UP000009046 UP000265180 UP000261380 UP000002852 UP000265000 UP000261560 UP000265200 UP000261440 UP000018467 UP000261400 UP000265020 UP000221080 UP000264800 UP000005207 UP000261500 UP000261480 UP000028760 UP000242638 UP000261640 UP000192220 UP000242188 UP000265100 UP000265160 UP000000437 UP000265040 UP000264820
Interpro
ProteinModelPortal
A0A2H1VMR1
A0A1E1WJU4
L7X1G4
D0AB91
A0A194QS01
A0A0L7LGR6
+ More
A0A212EP59 A0A067QYQ3 A0A146LJ53 A0A2J7PXZ6 A0A146LWA4 D6WW09 A0A2P8ZLG4 A0A1B6KTR6 A0A1B6EA14 A0A154PA95 W5JGP2 A0A182J889 A0A0M9A9K2 U4U402 N6SU96 A0A182M0H1 A0A182VTZ2 A0A182NBH5 A0A182R3W6 J9KG38 A0A023F0S3 A0A310SHW6 A0A2S2QP19 A0A224XFN9 A0A0T6AZL6 A0A1B0ETL7 A0A069DSR6 A0A0P4VIT5 A0A2R7WW04 A0A0L7RDP7 A0A026VZ81 A0A1Y1KI19 A0A1B6IS96 A0A1B6H5P9 A0A195CRW1 A0A151XAD2 A0A3M6UJD4 A0A195EDW5 A0A151I3U3 C3YR00 A0A2B4RSP0 A0A1A8UA31 A0A1A7ZUX4 A0A1A8CD96 A0A3B3HC86 A0A1S3JSC1 E0VAJ8 A0A3P9LXE9 A0A1A8F3T9 A0A3B5KTP5 A0A1A7XMY2 M3ZSJ8 A0A1A8IYQ6 A0A1A8PFK5 A0A3Q2PQP1 A0A3B3CCN6 A0A3P9JSC5 A0A3B4E6T0 A0A3B5RC92 A0A3B5L6R9 W5LKE6 A0A3S2MZ49 A0A3B4ZY20 A0A146QWA2 A0A3Q2CW62 A0A3Q2PP69 A0A315WBV3 A0A2D0QWW1 A0A3Q2ZNK3 I3JHL3 A0A3B3UWV6 A0A3B3YXE0 A0A087X8J9 A0A3P9P2A0 A0A3Q3MMN0 A0A3Q2QVN8 A0A3Q3M567 A0A147B0E9 A0A0S7EI22 A0A2I4C784 A0A146PQC1 Q9DGL7 Q6NWG2 A0A210QD94 A0A3P8Q9I1 A0A3P9D373 A0A3Q3MAF1 A0A0B7BH20 Q1L9C6 A0A3Q1HMM0 A0A3Q2XWN8
A0A212EP59 A0A067QYQ3 A0A146LJ53 A0A2J7PXZ6 A0A146LWA4 D6WW09 A0A2P8ZLG4 A0A1B6KTR6 A0A1B6EA14 A0A154PA95 W5JGP2 A0A182J889 A0A0M9A9K2 U4U402 N6SU96 A0A182M0H1 A0A182VTZ2 A0A182NBH5 A0A182R3W6 J9KG38 A0A023F0S3 A0A310SHW6 A0A2S2QP19 A0A224XFN9 A0A0T6AZL6 A0A1B0ETL7 A0A069DSR6 A0A0P4VIT5 A0A2R7WW04 A0A0L7RDP7 A0A026VZ81 A0A1Y1KI19 A0A1B6IS96 A0A1B6H5P9 A0A195CRW1 A0A151XAD2 A0A3M6UJD4 A0A195EDW5 A0A151I3U3 C3YR00 A0A2B4RSP0 A0A1A8UA31 A0A1A7ZUX4 A0A1A8CD96 A0A3B3HC86 A0A1S3JSC1 E0VAJ8 A0A3P9LXE9 A0A1A8F3T9 A0A3B5KTP5 A0A1A7XMY2 M3ZSJ8 A0A1A8IYQ6 A0A1A8PFK5 A0A3Q2PQP1 A0A3B3CCN6 A0A3P9JSC5 A0A3B4E6T0 A0A3B5RC92 A0A3B5L6R9 W5LKE6 A0A3S2MZ49 A0A3B4ZY20 A0A146QWA2 A0A3Q2CW62 A0A3Q2PP69 A0A315WBV3 A0A2D0QWW1 A0A3Q2ZNK3 I3JHL3 A0A3B3UWV6 A0A3B3YXE0 A0A087X8J9 A0A3P9P2A0 A0A3Q3MMN0 A0A3Q2QVN8 A0A3Q3M567 A0A147B0E9 A0A0S7EI22 A0A2I4C784 A0A146PQC1 Q9DGL7 Q6NWG2 A0A210QD94 A0A3P8Q9I1 A0A3P9D373 A0A3Q3MAF1 A0A0B7BH20 Q1L9C6 A0A3Q1HMM0 A0A3Q2XWN8
PDB
1V0D
E-value=5.51898e-34,
Score=360
Ontologies
GO
PANTHER
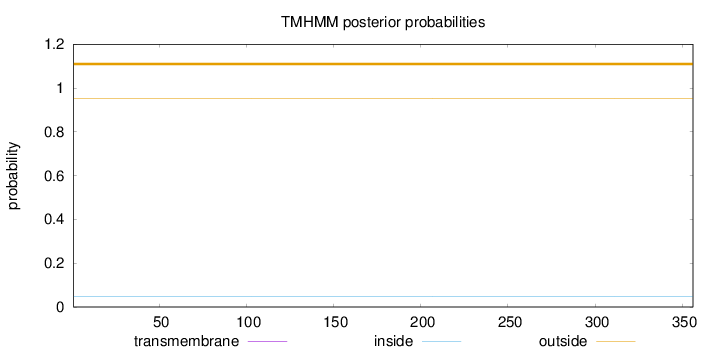
Topology
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000490000000000001
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.04825
outside
1 - 356
Population Genetic Test Statistics
Pi
8.313663
Theta
13.196514
Tajima's D
-1.27026
CLR
0.058256
CSRT
0.0884455777211139
Interpretation
Uncertain
