Gene
KWMTBOMO16528
Annotation
PREDICTED:_thymidylate_kinase-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.353
Sequence
CDS
ATGAAACGAGGTGCTTTAATAGTTATTGAAGGTATCGATAGAACAGGGAAGTCGACACAAGGGAAAATATTAGTTGAAAGCTTGAAAAAGCACAACATTGAAGCTGAATACAGAAATTTTCCTGCCCGTGAGACAGAAATTGGAAAAGTCATAAATAATTATCTATCTTCAAAGAATGAATTATCAGATGAAGCTATCCATCTACTATTTTCTGCGAACCGCTGGGAGCGAGCCCAAAATATAATAAAAACTTTAGAAGGAGGCACAACAATAGTGCTTGATAGATACTGTTACTCAGGTGTAGCCTTTTCAGCAGCAAAAGGTTTAGATCTGAACTGGTGCAAGTCTCCAGATGCTGGTTTGCCACAGCCAGACAAAGTTTTCTTTCTAACCATGCCTCTTGAAGCAGTTTTGAAAAGGAATGGTTTTGGGAACGAAAGATATGAAGTGGTAGATTTCCAAAAGAAAGTGTGGAATATTTATTCTAAATTGATAGAACCGAATTGGGACGTCCTGGATGCTAACAGAACAATGGACGCAATTCAAACAGATATTCTAAGCAAATCATTGGAGATCATAAAGGAGGCTGAAAATAGCCCCATAAAAAAAATATGGATTGAATCGTAA
Protein
MKRGALIVIEGIDRTGKSTQGKILVESLKKHNIEAEYRNFPARETEIGKVINNYLSSKNELSDEAIHLLFSANRWERAQNIIKTLEGGTTIVLDRYCYSGVAFSAAKGLDLNWCKSPDAGLPQPDKVFFLTMPLEAVLKRNGFGNERYEVVDFQKKVWNIYSKLIEPNWDVLDANRTMDAIQTDILSKSLEIIKEAENSPIKKIWIES
Summary
Uniprot
A0A2A4JCZ2
A0A2H1VL46
A0A0N1PFY9
A0A212EPA4
D0AB90
L7X3W4
+ More
A0A194QSK5 R4ZGJ3 A0A0D2WR91 A0A1B6EN76 A0A182WMH5 A0A2P6VHI3 A0A182VNW9 A0A1B6LBU9 A0A182X8H1 Q7PPZ1 A0A182LGS4 A0A1B6M0Z6 A0A0L0G1C5 A0A182HNS4 A0A0L0H7G7 A0A182K819 E1Z8Z0 A0A182YBY4 A0A182S709 A0A182M4R4 A0A182RP36 A0A1B0CYK9 E4YHT7 A0A2R5GGP8 E4X489 Q17LP5 A0A2P6TY72 A0A1S4EY68 C3ZEC2 V4B0U3 A0A182IQX1 A0A067REX9 A0A1D2NBB1 A0A182Q2G2 W8C5G7 A0A1B6EFI8 A0A182NV10 A0A0F8ANZ3 A0A1Y1XTZ2 A0A1Y1ICV4 N6TSA6 A0A3B3REA8 A0A336K3L0 A0A210PQL2 A0A2P6N7R7 F6SK91 A0A1L8DXV4 A0A182TXI2 B3SBN8 A0A1I8N896 U4UH17 A0A336L6X4 A0A1D2A8Z6 A0A2G4SJV5 A0A1X0QMY3 A0A367K2B2 A0A3Q3AGQ9 A0A369S0L9 B0W5K5 A0A0P4WLI0 T2MH14 A0A1Q3EVR0 A7S8M7 A0A3B3WT71 A0A3P9PIK3 A0A3B3U8P7 A0A087YL14 A0A023F7Z7 A0A1Y2C173 A0A0A1XG45 A0A2H3YH15 A0A3Q3RY18 A9TG14 A0A238F9H8 A0A1S3BYU7 A0A2A3EH77 A0A367J764 A0A0T6BHV6 A0A2H3YGM4 A0A151MJV9 A0A2H3YGM8 A0A0K8VVF7 A0A2K1JMM8 M3ZNH6 A0A3B5LIS9 A0A0S7H0S9 A0A1S3BY65 A0A367K0M1 A0A0A1ND87 A0A1Y2GN55 H2TXQ6 A0A2T0FLV0 A0A3Q3GTF3 V5GJC1
A0A194QSK5 R4ZGJ3 A0A0D2WR91 A0A1B6EN76 A0A182WMH5 A0A2P6VHI3 A0A182VNW9 A0A1B6LBU9 A0A182X8H1 Q7PPZ1 A0A182LGS4 A0A1B6M0Z6 A0A0L0G1C5 A0A182HNS4 A0A0L0H7G7 A0A182K819 E1Z8Z0 A0A182YBY4 A0A182S709 A0A182M4R4 A0A182RP36 A0A1B0CYK9 E4YHT7 A0A2R5GGP8 E4X489 Q17LP5 A0A2P6TY72 A0A1S4EY68 C3ZEC2 V4B0U3 A0A182IQX1 A0A067REX9 A0A1D2NBB1 A0A182Q2G2 W8C5G7 A0A1B6EFI8 A0A182NV10 A0A0F8ANZ3 A0A1Y1XTZ2 A0A1Y1ICV4 N6TSA6 A0A3B3REA8 A0A336K3L0 A0A210PQL2 A0A2P6N7R7 F6SK91 A0A1L8DXV4 A0A182TXI2 B3SBN8 A0A1I8N896 U4UH17 A0A336L6X4 A0A1D2A8Z6 A0A2G4SJV5 A0A1X0QMY3 A0A367K2B2 A0A3Q3AGQ9 A0A369S0L9 B0W5K5 A0A0P4WLI0 T2MH14 A0A1Q3EVR0 A7S8M7 A0A3B3WT71 A0A3P9PIK3 A0A3B3U8P7 A0A087YL14 A0A023F7Z7 A0A1Y2C173 A0A0A1XG45 A0A2H3YH15 A0A3Q3RY18 A9TG14 A0A238F9H8 A0A1S3BYU7 A0A2A3EH77 A0A367J764 A0A0T6BHV6 A0A2H3YGM4 A0A151MJV9 A0A2H3YGM8 A0A0K8VVF7 A0A2K1JMM8 M3ZNH6 A0A3B5LIS9 A0A0S7H0S9 A0A1S3BY65 A0A367K0M1 A0A0A1ND87 A0A1Y2GN55 H2TXQ6 A0A2T0FLV0 A0A3Q3GTF3 V5GJC1
Pubmed
26354079
22118469
23674305
23678178
29178410
12364791
+ More
20966253 20852019 25244985 21097902 17510324 18563158 23254933 24845553 27289101 24495485 25835551 24865297 23537049 29240929 28812685 29378020 12481130 15114417 18719581 25315136 27956601 29674435 30042472 24065732 17615350 25474469 25830018 23917264 18079367 22753475 22293439 29237241 23542700 21551351
20966253 20852019 25244985 21097902 17510324 18563158 23254933 24845553 27289101 24495485 25835551 24865297 23537049 29240929 28812685 29378020 12481130 15114417 18719581 25315136 27956601 29674435 30042472 24065732 17615350 25474469 25830018 23917264 18079367 22753475 22293439 29237241 23542700 21551351
EMBL
NWSH01001832
PCG69967.1
ODYU01003152
SOQ41543.1
KQ459232
KQ459127
+ More
KPJ02634.1 KPJ03781.1 AGBW02013517 OWR43287.1 FP102340 CBH09285.1 KC469893 AGC92695.1 KQ461155 KPJ08299.1 HF679134 CCU56373.1 KE346365 KJE93668.1 GECZ01030404 JAS39365.1 LHPF02000006 PSC73553.1 GEBQ01018826 GEBQ01010692 JAT21151.1 JAT29285.1 AAAB01008900 EAA09514.4 GEBQ01018403 GEBQ01010417 JAT21574.1 JAT29560.1 KQ241912 KNC82591.1 APCN01002756 KQ257465 KNC97162.1 GL433839 EFN57425.1 AXCM01019387 AJVK01001967 FN654580 CBY35059.1 BEYU01000065 GBG29775.1 FN653024 CBY23878.1 CH477213 EAT47657.1 LHPG02000004 PRW59003.1 GG666612 EEN49078.1 KB200639 ESP00866.1 KK852730 KDR17511.1 LJIJ01000107 ODN02539.1 AXCN02001010 GAMC01004884 JAC01672.1 GEDC01000601 JAS36697.1 KQ041311 KKF28478.1 MCFE01000465 ORX89231.1 DF237404 GAQ88740.1 APGK01056917 KB741277 ENN71201.1 UFQS01000086 UFQT01000086 SSW99077.1 SSX19459.1 NEDP02005555 OWF38767.1 MDYQ01000164 PRP79990.1 GFDF01002845 JAV11239.1 DS985265 EDV19916.1 KB632155 ERL89190.1 UFQS01001813 UFQT01001813 SSX12427.1 SSX31878.1 GDKF01002942 JAT75680.1 KZ303860 PHZ09041.1 KV922185 ORE01091.1 PJQM01002325 RCH96297.1 NOWV01000129 RDD39593.1 DS231843 EDS35488.1 GDRN01063553 GDRN01063552 GDRN01063551 GDRN01063550 GDRN01063549 JAI64965.1 HAAD01004978 CDG71210.1 GFDL01015650 JAV19395.1 DS469598 EDO39969.1 AYCK01022117 GBBI01001330 JAC17382.1 MCGR01000140 ORY40627.1 GBXI01003943 JAD10349.1 DS545112 EDQ57588.1 FMSP01000004 SCV69409.1 KZ288256 PBC30569.1 PJQL01002009 RCH85812.1 LJIG01000040 KRT86900.1 AKHW03006003 KYO24806.1 GDHF01009452 JAI42862.1 ABEU02000013 PNR42792.1 GBYX01446682 GBYX01446681 JAO34764.1 PJQL01000436 RCH95736.1 CDGI01000245 KV921263 CEI94785.1 ORE22844.1 MCFF01000022 ORZ13930.1 NDIQ01000022 PRT55973.1 GALX01006754 JAB61712.1
KPJ02634.1 KPJ03781.1 AGBW02013517 OWR43287.1 FP102340 CBH09285.1 KC469893 AGC92695.1 KQ461155 KPJ08299.1 HF679134 CCU56373.1 KE346365 KJE93668.1 GECZ01030404 JAS39365.1 LHPF02000006 PSC73553.1 GEBQ01018826 GEBQ01010692 JAT21151.1 JAT29285.1 AAAB01008900 EAA09514.4 GEBQ01018403 GEBQ01010417 JAT21574.1 JAT29560.1 KQ241912 KNC82591.1 APCN01002756 KQ257465 KNC97162.1 GL433839 EFN57425.1 AXCM01019387 AJVK01001967 FN654580 CBY35059.1 BEYU01000065 GBG29775.1 FN653024 CBY23878.1 CH477213 EAT47657.1 LHPG02000004 PRW59003.1 GG666612 EEN49078.1 KB200639 ESP00866.1 KK852730 KDR17511.1 LJIJ01000107 ODN02539.1 AXCN02001010 GAMC01004884 JAC01672.1 GEDC01000601 JAS36697.1 KQ041311 KKF28478.1 MCFE01000465 ORX89231.1 DF237404 GAQ88740.1 APGK01056917 KB741277 ENN71201.1 UFQS01000086 UFQT01000086 SSW99077.1 SSX19459.1 NEDP02005555 OWF38767.1 MDYQ01000164 PRP79990.1 GFDF01002845 JAV11239.1 DS985265 EDV19916.1 KB632155 ERL89190.1 UFQS01001813 UFQT01001813 SSX12427.1 SSX31878.1 GDKF01002942 JAT75680.1 KZ303860 PHZ09041.1 KV922185 ORE01091.1 PJQM01002325 RCH96297.1 NOWV01000129 RDD39593.1 DS231843 EDS35488.1 GDRN01063553 GDRN01063552 GDRN01063551 GDRN01063550 GDRN01063549 JAI64965.1 HAAD01004978 CDG71210.1 GFDL01015650 JAV19395.1 DS469598 EDO39969.1 AYCK01022117 GBBI01001330 JAC17382.1 MCGR01000140 ORY40627.1 GBXI01003943 JAD10349.1 DS545112 EDQ57588.1 FMSP01000004 SCV69409.1 KZ288256 PBC30569.1 PJQL01002009 RCH85812.1 LJIG01000040 KRT86900.1 AKHW03006003 KYO24806.1 GDHF01009452 JAI42862.1 ABEU02000013 PNR42792.1 GBYX01446682 GBYX01446681 JAO34764.1 PJQL01000436 RCH95736.1 CDGI01000245 KV921263 CEI94785.1 ORE22844.1 MCFF01000022 ORZ13930.1 NDIQ01000022 PRT55973.1 GALX01006754 JAB61712.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000014926
UP000008743
+ More
UP000075920 UP000239649 UP000075903 UP000076407 UP000007062 UP000075882 UP000054560 UP000075840 UP000053201 UP000075881 UP000008141 UP000076408 UP000075901 UP000075883 UP000075900 UP000092462 UP000008820 UP000239899 UP000001554 UP000030746 UP000075880 UP000027135 UP000094527 UP000075886 UP000075884 UP000193498 UP000054558 UP000019118 UP000261540 UP000242188 UP000241769 UP000008144 UP000075902 UP000009022 UP000095301 UP000030742 UP000242254 UP000242414 UP000253551 UP000264800 UP000253843 UP000002320 UP000001593 UP000261480 UP000242638 UP000261500 UP000028760 UP000193467 UP000228380 UP000261640 UP000198372 UP000089565 UP000242457 UP000252139 UP000050525 UP000006727 UP000002852 UP000261380 UP000038169 UP000242381 UP000193648 UP000005226 UP000238350 UP000261660
UP000075920 UP000239649 UP000075903 UP000076407 UP000007062 UP000075882 UP000054560 UP000075840 UP000053201 UP000075881 UP000008141 UP000076408 UP000075901 UP000075883 UP000075900 UP000092462 UP000008820 UP000239899 UP000001554 UP000030746 UP000075880 UP000027135 UP000094527 UP000075886 UP000075884 UP000193498 UP000054558 UP000019118 UP000261540 UP000242188 UP000241769 UP000008144 UP000075902 UP000009022 UP000095301 UP000030742 UP000242254 UP000242414 UP000253551 UP000264800 UP000253843 UP000002320 UP000001593 UP000261480 UP000242638 UP000261500 UP000028760 UP000193467 UP000228380 UP000261640 UP000198372 UP000089565 UP000242457 UP000252139 UP000050525 UP000006727 UP000002852 UP000261380 UP000038169 UP000242381 UP000193648 UP000005226 UP000238350 UP000261660
Pfam
PF02223 Thymidylate_kin
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JCZ2
A0A2H1VL46
A0A0N1PFY9
A0A212EPA4
D0AB90
L7X3W4
+ More
A0A194QSK5 R4ZGJ3 A0A0D2WR91 A0A1B6EN76 A0A182WMH5 A0A2P6VHI3 A0A182VNW9 A0A1B6LBU9 A0A182X8H1 Q7PPZ1 A0A182LGS4 A0A1B6M0Z6 A0A0L0G1C5 A0A182HNS4 A0A0L0H7G7 A0A182K819 E1Z8Z0 A0A182YBY4 A0A182S709 A0A182M4R4 A0A182RP36 A0A1B0CYK9 E4YHT7 A0A2R5GGP8 E4X489 Q17LP5 A0A2P6TY72 A0A1S4EY68 C3ZEC2 V4B0U3 A0A182IQX1 A0A067REX9 A0A1D2NBB1 A0A182Q2G2 W8C5G7 A0A1B6EFI8 A0A182NV10 A0A0F8ANZ3 A0A1Y1XTZ2 A0A1Y1ICV4 N6TSA6 A0A3B3REA8 A0A336K3L0 A0A210PQL2 A0A2P6N7R7 F6SK91 A0A1L8DXV4 A0A182TXI2 B3SBN8 A0A1I8N896 U4UH17 A0A336L6X4 A0A1D2A8Z6 A0A2G4SJV5 A0A1X0QMY3 A0A367K2B2 A0A3Q3AGQ9 A0A369S0L9 B0W5K5 A0A0P4WLI0 T2MH14 A0A1Q3EVR0 A7S8M7 A0A3B3WT71 A0A3P9PIK3 A0A3B3U8P7 A0A087YL14 A0A023F7Z7 A0A1Y2C173 A0A0A1XG45 A0A2H3YH15 A0A3Q3RY18 A9TG14 A0A238F9H8 A0A1S3BYU7 A0A2A3EH77 A0A367J764 A0A0T6BHV6 A0A2H3YGM4 A0A151MJV9 A0A2H3YGM8 A0A0K8VVF7 A0A2K1JMM8 M3ZNH6 A0A3B5LIS9 A0A0S7H0S9 A0A1S3BY65 A0A367K0M1 A0A0A1ND87 A0A1Y2GN55 H2TXQ6 A0A2T0FLV0 A0A3Q3GTF3 V5GJC1
A0A194QSK5 R4ZGJ3 A0A0D2WR91 A0A1B6EN76 A0A182WMH5 A0A2P6VHI3 A0A182VNW9 A0A1B6LBU9 A0A182X8H1 Q7PPZ1 A0A182LGS4 A0A1B6M0Z6 A0A0L0G1C5 A0A182HNS4 A0A0L0H7G7 A0A182K819 E1Z8Z0 A0A182YBY4 A0A182S709 A0A182M4R4 A0A182RP36 A0A1B0CYK9 E4YHT7 A0A2R5GGP8 E4X489 Q17LP5 A0A2P6TY72 A0A1S4EY68 C3ZEC2 V4B0U3 A0A182IQX1 A0A067REX9 A0A1D2NBB1 A0A182Q2G2 W8C5G7 A0A1B6EFI8 A0A182NV10 A0A0F8ANZ3 A0A1Y1XTZ2 A0A1Y1ICV4 N6TSA6 A0A3B3REA8 A0A336K3L0 A0A210PQL2 A0A2P6N7R7 F6SK91 A0A1L8DXV4 A0A182TXI2 B3SBN8 A0A1I8N896 U4UH17 A0A336L6X4 A0A1D2A8Z6 A0A2G4SJV5 A0A1X0QMY3 A0A367K2B2 A0A3Q3AGQ9 A0A369S0L9 B0W5K5 A0A0P4WLI0 T2MH14 A0A1Q3EVR0 A7S8M7 A0A3B3WT71 A0A3P9PIK3 A0A3B3U8P7 A0A087YL14 A0A023F7Z7 A0A1Y2C173 A0A0A1XG45 A0A2H3YH15 A0A3Q3RY18 A9TG14 A0A238F9H8 A0A1S3BYU7 A0A2A3EH77 A0A367J764 A0A0T6BHV6 A0A2H3YGM4 A0A151MJV9 A0A2H3YGM8 A0A0K8VVF7 A0A2K1JMM8 M3ZNH6 A0A3B5LIS9 A0A0S7H0S9 A0A1S3BY65 A0A367K0M1 A0A0A1ND87 A0A1Y2GN55 H2TXQ6 A0A2T0FLV0 A0A3Q3GTF3 V5GJC1
PDB
2XX3
E-value=2.37377e-51,
Score=507
Ontologies
PATHWAY
GO
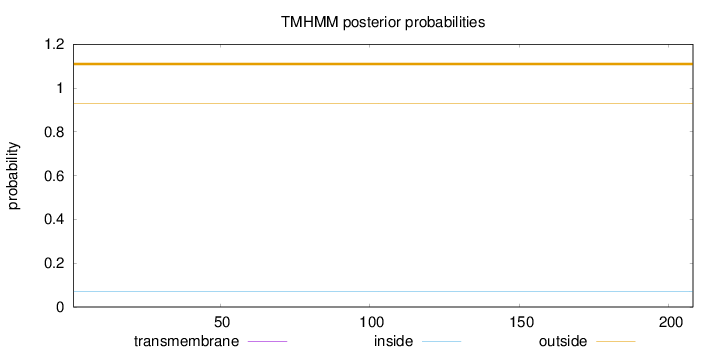
Topology
Length:
208
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00129
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07019
outside
1 - 208
Population Genetic Test Statistics
Pi
4.306065
Theta
14.069207
Tajima's D
-2.000378
CLR
0
CSRT
0.0138493075346233
Interpretation
Uncertain
