Gene
KWMTBOMO16523
Annotation
HM00048_[Heliconius_melpomene]
Location in the cell
Mitochondrial Reliability : 2.701
Sequence
CDS
ATGTCTGGAGTAAAGCAAGTACGAAATAAAAAGCTTCTCCCGGATTTGCGTAAAGAAGGCGAACTCACTAAAGAAATAATATTATCTACGAAGGAGAAACACGGAGTTCCTGTGACGTCTAGACTTTTTTCTCATCATACCTTAGCGAGCGTTAGAAAACTAAGCTTTTATTTTCCTTATAATTTGCCAGACGACAGCCTGGACTTCATTTTAACGTCCAAATACAATCACTCGACGGAGCAATTTGCGGATAAAGTCGACGTGTTCCTGCAGCCAGAGACGATCGGTTGCGAGACCTGGCGACGACTTCGAAACACTTGTGATAAATTACCAAGCAGTGGAGTCCCAATGGTGAGTTTTCACTTGAAAAAGAAGGCACTAGCTATTTCTAAATATATCTTTTAG
Protein
MSGVKQVRNKKLLPDLRKEGELTKEIILSTKEKHGVPVTSRLFSHHTLASVRKLSFYFPYNLPDDSLDFILTSKYNHSTEQFADKVDVFLQPETIGCETWRRLRNTCDKLPSSGVPMVSFHLKKKALAISKYIF
Summary
Uniprot
L7X1H0
D0AB96
A0A212EPA0
A0A194QRZ7
A0A2A4K2E0
A0A194QCD0
+ More
A0A0L7KSZ0 D6WYE4 A0A1W4WIZ9 N6TPS1 U4U0U1 A0A026WZA8 B0WIP4 A0A084WU35 A0A1Q3FSS7 A0A1Q3FSQ1 A0A1J1J5S2 A0A182Y0N1 A0A182T8S6 A0A182M7F0 A0A1B0CZZ7 A0A182P4Q3 Q5TP55 A0A182G3M9 A0A0J7KXJ1 Q16T95 A0A195B2B1 F4WW09 A0A2C9GR35 E2B0S6 A0A023EH39 A0A195E652 E2B5P1 A0A336LSQ2 A0A154NWC6 A0A158NLI7 A0A2A3EPT7 A0A151IMG0 A0A088AG61 A0A182K0H1 A0A232EKP7 A0A1A9XW07 A0A1A9UH86 K7J077 A0A310SAA0 A0A195EYY2 A0A0L7QLL7 A0A1B0FAT9 A0A0A9YHD8 A0A0M8ZXP2 A0A1A9WZQ7 A0A1I8PL85 A0A151XCW4 B4JCQ2 A0A2C9JH05 A0A2C9JH08 A0A034WHL6 A0A0L0C5I6 A0A0K8VL56 A0A1I8MZM8 B4G8T2 A0A0M5J9I1 Q29MS5 A0A3B0JFF9 W8BGP7 B4KJ39 A0A2R7WXW5 A0A1B6JAB9 A0A1B6BYY3 A0A1W4V339 B4P215 Q9VK88 B4IE90 B4Q3T2 B4MWL8 B3N3U6 A0A3M6THZ8 B3MNU1
A0A0L7KSZ0 D6WYE4 A0A1W4WIZ9 N6TPS1 U4U0U1 A0A026WZA8 B0WIP4 A0A084WU35 A0A1Q3FSS7 A0A1Q3FSQ1 A0A1J1J5S2 A0A182Y0N1 A0A182T8S6 A0A182M7F0 A0A1B0CZZ7 A0A182P4Q3 Q5TP55 A0A182G3M9 A0A0J7KXJ1 Q16T95 A0A195B2B1 F4WW09 A0A2C9GR35 E2B0S6 A0A023EH39 A0A195E652 E2B5P1 A0A336LSQ2 A0A154NWC6 A0A158NLI7 A0A2A3EPT7 A0A151IMG0 A0A088AG61 A0A182K0H1 A0A232EKP7 A0A1A9XW07 A0A1A9UH86 K7J077 A0A310SAA0 A0A195EYY2 A0A0L7QLL7 A0A1B0FAT9 A0A0A9YHD8 A0A0M8ZXP2 A0A1A9WZQ7 A0A1I8PL85 A0A151XCW4 B4JCQ2 A0A2C9JH05 A0A2C9JH08 A0A034WHL6 A0A0L0C5I6 A0A0K8VL56 A0A1I8MZM8 B4G8T2 A0A0M5J9I1 Q29MS5 A0A3B0JFF9 W8BGP7 B4KJ39 A0A2R7WXW5 A0A1B6JAB9 A0A1B6BYY3 A0A1W4V339 B4P215 Q9VK88 B4IE90 B4Q3T2 B4MWL8 B3N3U6 A0A3M6THZ8 B3MNU1
Pubmed
23674305
22118469
26354079
26227816
18362917
19820115
+ More
23537049 24508170 30249741 24438588 25244985 12364791 14747013 17210077 26483478 17510324 21719571 20798317 24945155 21347285 28648823 20075255 25401762 17994087 15562597 25348373 26108605 25315136 15632085 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 30382153
23537049 24508170 30249741 24438588 25244985 12364791 14747013 17210077 26483478 17510324 21719571 20798317 24945155 21347285 28648823 20075255 25401762 17994087 15562597 25348373 26108605 25315136 15632085 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 30382153
EMBL
KC469893
AGC92701.2
FP102340
CBH09291.1
AGBW02013517
OWR43281.1
+ More
KQ461155 KPJ08293.1 NWSH01000227 PCG78219.1 KQ459232 KPJ02640.1 JTDY01005985 KOB66392.1 KQ971362 EFA07888.1 APGK01057000 APGK01057001 KB741277 ENN71240.1 KB631850 ERL86702.1 KK107063 QOIP01000005 EZA61076.1 RLU22957.1 DS231951 EDS28621.1 ATLV01026995 KE525421 KFB53729.1 GFDL01004418 JAV30627.1 GFDL01004416 JAV30629.1 CVRI01000073 CRL07755.1 AXCM01001134 AJVK01009786 AAAB01008980 EAL39185.1 JXUM01141868 KQ569321 KXJ68665.1 LBMM01002126 KMQ95242.1 CH477657 EAT37695.1 KQ976662 KYM78339.1 GL888404 EGI61598.1 APCN01001775 GL444666 EFN60748.1 GAPW01004915 JAC08683.1 KQ979579 KYN20648.1 GL445864 EFN89010.1 UFQT01000093 SSX19609.1 KQ434773 KZC03995.1 ADTU01019457 KZ288197 PBC33710.1 KQ977054 KYN06081.1 NNAY01003762 OXU18888.1 KQ762367 OAD55914.1 KQ981914 KYN33089.1 KQ414914 KOC59522.1 CCAG010023607 GBHO01013091 GBRD01004593 JAG30513.1 JAG61228.1 KQ435801 KOX73240.1 KQ982314 KYQ58098.1 CH916368 EDW04216.1 GAKP01005719 GAKP01005717 GAKP01005715 JAC53233.1 JRES01000869 KNC27658.1 GDHF01029370 GDHF01013044 JAI22944.1 JAI39270.1 CH479180 EDW28762.1 CP012523 ALC40044.1 CH379060 EAL33618.2 OUUW01000004 SPP78952.1 GAMC01008638 GAMC01008636 JAB97917.1 CH933807 EDW11401.1 KK855922 PTY24376.1 GECU01011580 JAS96126.1 GEDC01030815 JAS06483.1 CM000157 EDW88186.1 AE014134 AY118683 AAF53190.2 AAM50543.1 CH480831 EDW45917.1 CM000361 CM002910 EDX04807.1 KMY89919.1 CH963857 EDW76507.1 CH954177 EDV58798.1 RCHS01003527 RMX41047.1 CH902620 EDV32128.1
KQ461155 KPJ08293.1 NWSH01000227 PCG78219.1 KQ459232 KPJ02640.1 JTDY01005985 KOB66392.1 KQ971362 EFA07888.1 APGK01057000 APGK01057001 KB741277 ENN71240.1 KB631850 ERL86702.1 KK107063 QOIP01000005 EZA61076.1 RLU22957.1 DS231951 EDS28621.1 ATLV01026995 KE525421 KFB53729.1 GFDL01004418 JAV30627.1 GFDL01004416 JAV30629.1 CVRI01000073 CRL07755.1 AXCM01001134 AJVK01009786 AAAB01008980 EAL39185.1 JXUM01141868 KQ569321 KXJ68665.1 LBMM01002126 KMQ95242.1 CH477657 EAT37695.1 KQ976662 KYM78339.1 GL888404 EGI61598.1 APCN01001775 GL444666 EFN60748.1 GAPW01004915 JAC08683.1 KQ979579 KYN20648.1 GL445864 EFN89010.1 UFQT01000093 SSX19609.1 KQ434773 KZC03995.1 ADTU01019457 KZ288197 PBC33710.1 KQ977054 KYN06081.1 NNAY01003762 OXU18888.1 KQ762367 OAD55914.1 KQ981914 KYN33089.1 KQ414914 KOC59522.1 CCAG010023607 GBHO01013091 GBRD01004593 JAG30513.1 JAG61228.1 KQ435801 KOX73240.1 KQ982314 KYQ58098.1 CH916368 EDW04216.1 GAKP01005719 GAKP01005717 GAKP01005715 JAC53233.1 JRES01000869 KNC27658.1 GDHF01029370 GDHF01013044 JAI22944.1 JAI39270.1 CH479180 EDW28762.1 CP012523 ALC40044.1 CH379060 EAL33618.2 OUUW01000004 SPP78952.1 GAMC01008638 GAMC01008636 JAB97917.1 CH933807 EDW11401.1 KK855922 PTY24376.1 GECU01011580 JAS96126.1 GEDC01030815 JAS06483.1 CM000157 EDW88186.1 AE014134 AY118683 AAF53190.2 AAM50543.1 CH480831 EDW45917.1 CM000361 CM002910 EDX04807.1 KMY89919.1 CH963857 EDW76507.1 CH954177 EDV58798.1 RCHS01003527 RMX41047.1 CH902620 EDV32128.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000053268
UP000037510
UP000007266
+ More
UP000192223 UP000019118 UP000030742 UP000053097 UP000279307 UP000002320 UP000030765 UP000183832 UP000076408 UP000075901 UP000075883 UP000092462 UP000075885 UP000007062 UP000069940 UP000249989 UP000036403 UP000008820 UP000078540 UP000007755 UP000075840 UP000000311 UP000078492 UP000008237 UP000076502 UP000005205 UP000242457 UP000078542 UP000005203 UP000075881 UP000215335 UP000092443 UP000078200 UP000002358 UP000078541 UP000053825 UP000092444 UP000053105 UP000091820 UP000095300 UP000075809 UP000001070 UP000076420 UP000037069 UP000095301 UP000008744 UP000092553 UP000001819 UP000268350 UP000009192 UP000192221 UP000002282 UP000000803 UP000001292 UP000000304 UP000007798 UP000008711 UP000275408 UP000007801
UP000192223 UP000019118 UP000030742 UP000053097 UP000279307 UP000002320 UP000030765 UP000183832 UP000076408 UP000075901 UP000075883 UP000092462 UP000075885 UP000007062 UP000069940 UP000249989 UP000036403 UP000008820 UP000078540 UP000007755 UP000075840 UP000000311 UP000078492 UP000008237 UP000076502 UP000005205 UP000242457 UP000078542 UP000005203 UP000075881 UP000215335 UP000092443 UP000078200 UP000002358 UP000078541 UP000053825 UP000092444 UP000053105 UP000091820 UP000095300 UP000075809 UP000001070 UP000076420 UP000037069 UP000095301 UP000008744 UP000092553 UP000001819 UP000268350 UP000009192 UP000192221 UP000002282 UP000000803 UP000001292 UP000000304 UP000007798 UP000008711 UP000275408 UP000007801
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
L7X1H0
D0AB96
A0A212EPA0
A0A194QRZ7
A0A2A4K2E0
A0A194QCD0
+ More
A0A0L7KSZ0 D6WYE4 A0A1W4WIZ9 N6TPS1 U4U0U1 A0A026WZA8 B0WIP4 A0A084WU35 A0A1Q3FSS7 A0A1Q3FSQ1 A0A1J1J5S2 A0A182Y0N1 A0A182T8S6 A0A182M7F0 A0A1B0CZZ7 A0A182P4Q3 Q5TP55 A0A182G3M9 A0A0J7KXJ1 Q16T95 A0A195B2B1 F4WW09 A0A2C9GR35 E2B0S6 A0A023EH39 A0A195E652 E2B5P1 A0A336LSQ2 A0A154NWC6 A0A158NLI7 A0A2A3EPT7 A0A151IMG0 A0A088AG61 A0A182K0H1 A0A232EKP7 A0A1A9XW07 A0A1A9UH86 K7J077 A0A310SAA0 A0A195EYY2 A0A0L7QLL7 A0A1B0FAT9 A0A0A9YHD8 A0A0M8ZXP2 A0A1A9WZQ7 A0A1I8PL85 A0A151XCW4 B4JCQ2 A0A2C9JH05 A0A2C9JH08 A0A034WHL6 A0A0L0C5I6 A0A0K8VL56 A0A1I8MZM8 B4G8T2 A0A0M5J9I1 Q29MS5 A0A3B0JFF9 W8BGP7 B4KJ39 A0A2R7WXW5 A0A1B6JAB9 A0A1B6BYY3 A0A1W4V339 B4P215 Q9VK88 B4IE90 B4Q3T2 B4MWL8 B3N3U6 A0A3M6THZ8 B3MNU1
A0A0L7KSZ0 D6WYE4 A0A1W4WIZ9 N6TPS1 U4U0U1 A0A026WZA8 B0WIP4 A0A084WU35 A0A1Q3FSS7 A0A1Q3FSQ1 A0A1J1J5S2 A0A182Y0N1 A0A182T8S6 A0A182M7F0 A0A1B0CZZ7 A0A182P4Q3 Q5TP55 A0A182G3M9 A0A0J7KXJ1 Q16T95 A0A195B2B1 F4WW09 A0A2C9GR35 E2B0S6 A0A023EH39 A0A195E652 E2B5P1 A0A336LSQ2 A0A154NWC6 A0A158NLI7 A0A2A3EPT7 A0A151IMG0 A0A088AG61 A0A182K0H1 A0A232EKP7 A0A1A9XW07 A0A1A9UH86 K7J077 A0A310SAA0 A0A195EYY2 A0A0L7QLL7 A0A1B0FAT9 A0A0A9YHD8 A0A0M8ZXP2 A0A1A9WZQ7 A0A1I8PL85 A0A151XCW4 B4JCQ2 A0A2C9JH05 A0A2C9JH08 A0A034WHL6 A0A0L0C5I6 A0A0K8VL56 A0A1I8MZM8 B4G8T2 A0A0M5J9I1 Q29MS5 A0A3B0JFF9 W8BGP7 B4KJ39 A0A2R7WXW5 A0A1B6JAB9 A0A1B6BYY3 A0A1W4V339 B4P215 Q9VK88 B4IE90 B4Q3T2 B4MWL8 B3N3U6 A0A3M6THZ8 B3MNU1
Ontologies
GO
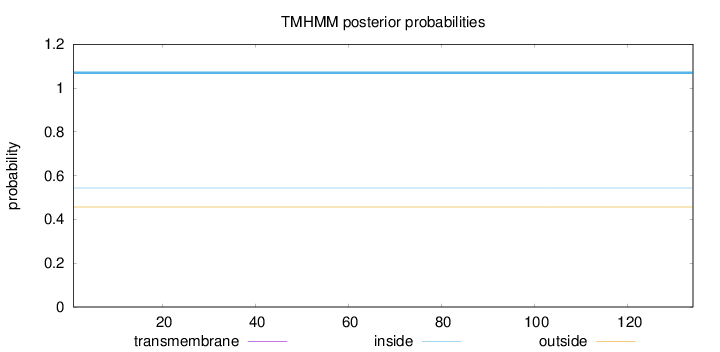
Topology
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00168
Exp number, first 60 AAs:
0.00091
Total prob of N-in:
0.54379
inside
1 - 134
Population Genetic Test Statistics
Pi
28.404212
Theta
33.289911
Tajima's D
-0.438988
CLR
0.184011
CSRT
0.255437228138593
Interpretation
Uncertain
