Gene
KWMTBOMO16521
Pre Gene Modal
BGIBMGA012137
Annotation
PREDICTED:_major_facilitator_superfamily_domain-containing_protein_6-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.305
Sequence
CDS
ATGAAGATTAACAAAAGCTTGCTGCCTGTAAAGGCGCATTTCTTCTTTTTCTTCGCCGCCCTGGGGCCGCTGTTGCCGCAAATGAATGTGTTCGGGCGGCAACTGGGAGTCACGCCCGCAGCGATGGGAATCGTGACGGCGATTCTGCCCCTGCTGTGGGCGACTGCGAAGCCACTCTTCGGCTACGTTGTAGATTATTGGCCGGCGCACAGGAAGCTAGTATTCATGCTGCTAATATCGGTTATGACCGGCTCGTACTGTTGCCTCTGGTTCTTGCCCACCCCGGAAACCGTCGAGCCGCCCCTCCTGGAATACGTTTATAAACTGGAAACCGTTGATTTGAGACGAAACGATCAGTCCCTGAACCTAGAAAAGTACACGTGCCACTGGAATTGTACTCCGGGAACGTTCGATGTGTACCTGCGGAACACGACCCTTATCGATACAGCATACTTGGACAGTGACGTCACGAATTCGACCTGTCTGACCCTGAACATGGACTTAAGCGAAGTCCGAGACGGGCAAGTGAAGTGCACGCCCAGAGACGGCTGCGATTTGTTGTGCTACGAAAAGGGATTGAATCGAGTCAAAAGAGAGATATCAGGTTCCACTAAGAATTTGGTTGACGTTAACGAGAAGATCTCTGGTGATTCTTCTGTTGAAACGGGGGGAGGAGGTAGCGTCTATGTGAGTCTTACGTTTTGGGCGTTCGTGGTTCTGATGAGTCTGGGTACAGTGGCGTTTAATGTGGCCAATTGTATCGGTGACGCCGTGTGCTTCGACGTGCTTGGTCCAGATCGAGGATCGAAATACGGTGCCCAACGAGCTTGGGGCACTATCGGATATGGGGTGACGGCCCTGCTGGGAGGTTGCCTGGTGGATATGGTATCTGATGGCAAGTACAAGGACTACACGCCGGCGTTCGTGATAGCCCTAATTGCGACCGGCATCGATCTTTATAGCTGCAGAGCGTTGACGCTACCCTCATTATCGAGCCCCACCGATTCCGGTGCGGCATTACGTTCTGTACTGCGGATACCGAAGGTGATTGTGTTCATAGTGTTCGCCGTACTGGCGGGCACATTCGACAGCTTCATAATCTATTACATGTTCTGGTTCCTGGAAGAATTGGCCGAGGAAACCGGTTCGATTGCTACGATAAAGCTCATCGAGGGCGTGGTGGTAGCCGCGCAGAGCTTCGTCGGGGAGCTTCTGTTCTTTTTCTGTTCAGGTAAAATAATAAAGCGCTGGGGCTACGGAACGACGCTGACGTTTTCATTGTTCTGTTACGGAATTAGAATGGCGTTGATATCGTTCATTCGGAATCCGTGGCAATTGGTGTTGATAGAGGGGATTATGCAAGGCCCGACATATGCCCTTTGCTACGCTACTATAGTCGGGTACGCGGCTCATGTGGCCCCGGACGGTTACTCGGCGACTGTCCAGGGCATAGTGGCCGGCATGGACGACGGCGTGGGTTTCGCCCTGGGATCGCTGCTAGGCGGACAACTGTACGGTTCGATGGGCGGTCGATATTCGTTCCGAGTGTTCAGCGTATTGGCGGTGATCGCGGCCGCCCTGCACGCCGCAATGTTCTTGGAATTCGATCGGGCTGCAGATTCCTGTTGGACGAATGACGATGGTGATAAGCCGAAGGTGACTCGGTTCGTTGGTAGACGCCAATCAGAAGTGGATGAAGACTGTGATGATTATAATTATGATGATGTCGTAACGCCTGATGATTTAGAGGAATACCAGGATTCAGACACGCGAGATTGCGATGAAAACATACAAATAGAAACGGTATATTGTTATGAAGACGATTCCACTTGTTATGTTGATAATAATAATGATGATGATGATTGA
Protein
MKINKSLLPVKAHFFFFFAALGPLLPQMNVFGRQLGVTPAAMGIVTAILPLLWATAKPLFGYVVDYWPAHRKLVFMLLISVMTGSYCCLWFLPTPETVEPPLLEYVYKLETVDLRRNDQSLNLEKYTCHWNCTPGTFDVYLRNTTLIDTAYLDSDVTNSTCLTLNMDLSEVRDGQVKCTPRDGCDLLCYEKGLNRVKREISGSTKNLVDVNEKISGDSSVETGGGGSVYVSLTFWAFVVLMSLGTVAFNVANCIGDAVCFDVLGPDRGSKYGAQRAWGTIGYGVTALLGGCLVDMVSDGKYKDYTPAFVIALIATGIDLYSCRALTLPSLSSPTDSGAALRSVLRIPKVIVFIVFAVLAGTFDSFIIYYMFWFLEELAEETGSIATIKLIEGVVVAAQSFVGELLFFFCSGKIIKRWGYGTTLTFSLFCYGIRMALISFIRNPWQLVLIEGIMQGPTYALCYATIVGYAAHVAPDGYSATVQGIVAGMDDGVGFALGSLLGGQLYGSMGGRYSFRVFSVLAVIAAALHAAMFLEFDRAADSCWTNDDGDKPKVTRFVGRRQSEVDEDCDDYNYDDVVTPDDLEEYQDSDTRDCDENIQIETVYCYEDDSTCYVDNNNDDDD
Summary
Uniprot
H9JRH7
A0A2H1VJP3
A0A0N1I937
A0A3S2PB24
A0A151ILP2
A0A151WEV2
+ More
A0A088A4G5 A0A2A3EEV4 A0A195FKJ5 A0A067QRS4 F4WR41 A0A195BBS0 A0A151JNH3 A0A158NVH6 E2C1J9 A0A2J7RH72 A0A2J7RH69 A0A1B0C8B2 A0A1J1IIW4 A0A154PKN9 A0A1L8DEF4 A0A1L8DEL6 A0A0K8UKI2 A0A1I8MUF3 A0A2P8Y492 A0A0K8UGX1 A0A0K8UTZ9 A0A034VX86 W8APK8 A0A0L0CKC8 T1P8J8 A0A3B0JJV7 A0A1B0GBJ0 A0A1B0BXB7 A0A1A9XEC7 B4MKD9 A0A1A9VE41 E9IWP5 B3NPV0 A0A1L8DDZ6 A0A0A1XC57 B4QHH7 A0A1I8NSP7 Q8MRB2 A0A1L8DE28 B4HST6 A0A1W4VCP7 B4LKY1 A0A1W4WDG2 B4GI85 B3MH64 B4J5J3 B5DZ61 K7J5C1 B4KPS0 A0A0M5IXW4 B4P6B6 D6WCF4 A0A1W4WEF1 A0A0N0BFA9 A0A224XI40 A0A1Y1LZ67 A0A1Y1LRF2 A0A1Y1LTU5 A0A1B6CJ75 U4U7K6 N6TWM6 F4WE94 A0A0T6BCR0 A0A2R7W644 A0A1W4X5L7 A0A1W4X6V3 A0A151ILN9 A0A151ILP7 A0A195FG52 A0A139WN88 A0A195C8R6 A0A158NUB4 A0A146LKJ3 A0A151WES1 A0A151I5A5 A0A151ILR1 A0A195FJN8 A0A195BBT0 A0A195EJZ4 A0A158NVI0 A0A195FH24 A0A195FLK8 A0A158NJJ1
A0A088A4G5 A0A2A3EEV4 A0A195FKJ5 A0A067QRS4 F4WR41 A0A195BBS0 A0A151JNH3 A0A158NVH6 E2C1J9 A0A2J7RH72 A0A2J7RH69 A0A1B0C8B2 A0A1J1IIW4 A0A154PKN9 A0A1L8DEF4 A0A1L8DEL6 A0A0K8UKI2 A0A1I8MUF3 A0A2P8Y492 A0A0K8UGX1 A0A0K8UTZ9 A0A034VX86 W8APK8 A0A0L0CKC8 T1P8J8 A0A3B0JJV7 A0A1B0GBJ0 A0A1B0BXB7 A0A1A9XEC7 B4MKD9 A0A1A9VE41 E9IWP5 B3NPV0 A0A1L8DDZ6 A0A0A1XC57 B4QHH7 A0A1I8NSP7 Q8MRB2 A0A1L8DE28 B4HST6 A0A1W4VCP7 B4LKY1 A0A1W4WDG2 B4GI85 B3MH64 B4J5J3 B5DZ61 K7J5C1 B4KPS0 A0A0M5IXW4 B4P6B6 D6WCF4 A0A1W4WEF1 A0A0N0BFA9 A0A224XI40 A0A1Y1LZ67 A0A1Y1LRF2 A0A1Y1LTU5 A0A1B6CJ75 U4U7K6 N6TWM6 F4WE94 A0A0T6BCR0 A0A2R7W644 A0A1W4X5L7 A0A1W4X6V3 A0A151ILN9 A0A151ILP7 A0A195FG52 A0A139WN88 A0A195C8R6 A0A158NUB4 A0A146LKJ3 A0A151WES1 A0A151I5A5 A0A151ILR1 A0A195FJN8 A0A195BBT0 A0A195EJZ4 A0A158NVI0 A0A195FH24 A0A195FLK8 A0A158NJJ1
Pubmed
19121390
26354079
24845553
21719571
21347285
20798317
+ More
25315136 29403074 25348373 24495485 26108605 17994087 21282665 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 23185243 20075255 17550304 18362917 19820115 28004739 23537049 26823975
25315136 29403074 25348373 24495485 26108605 17994087 21282665 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 23185243 20075255 17550304 18362917 19820115 28004739 23537049 26823975
EMBL
BABH01031908
BABH01031909
ODYU01002673
SOQ40474.1
KQ460648
KPJ13153.1
+ More
RSAL01000132 RVE46368.1 KQ977104 KYN05808.1 KQ983238 KYQ46342.1 KZ288266 PBC30248.1 KQ981512 KYN40876.1 KK853097 KDR11427.1 GL888284 EGI63268.1 KQ976532 KYM81655.1 KQ978884 KYN27677.1 ADTU01027161 GL451937 EFN78175.1 NEVH01003746 PNF40181.1 PNF40182.1 AJWK01000310 AJWK01000311 AJWK01000312 AJWK01000313 AJWK01000314 CVRI01000047 CRK98393.1 KQ434948 KZC12393.1 GFDF01009258 JAV04826.1 GFDF01009259 JAV04825.1 GDHF01025120 JAI27194.1 PYGN01000948 PSN39063.1 GDHF01026511 GDHF01003173 JAI25803.1 JAI49141.1 GDHF01022509 JAI29805.1 GAKP01011016 JAC47936.1 GAMC01020117 JAB86438.1 JRES01000264 KNC32853.1 KA645081 AFP59710.1 OUUW01000001 SPP72891.1 CCAG010014016 JXJN01022164 JXJN01022165 CH963846 EDW72578.1 GL766597 EFZ15023.1 CH954179 EDV55797.1 GFDF01009385 JAV04699.1 GBXI01013198 GBXI01012822 GBXI01007590 GBXI01005757 JAD01094.1 JAD01470.1 JAD06702.1 JAD08535.1 CM000362 CM002911 EDX07322.1 KMY94211.1 AE013599 AY121685 AAF58040.2 AAM52012.1 GFDF01009386 JAV04698.1 CH480816 EDW48100.1 CH940648 EDW60785.1 KRF79615.1 KRF79616.1 KRF79617.1 KRF79618.1 CH479183 EDW36205.1 CH902619 EDV35823.1 KPU75843.1 KPU75844.1 KPU75845.1 KPU75846.1 CH916367 EDW00756.1 CM000071 EDY69160.1 KRT02513.1 AAZX01004900 CH933808 EDW10197.1 KRG05084.1 KRG05085.1 KRG05086.1 CP012524 ALC42236.1 CM000158 EDW91966.1 KRK00222.1 KRK00223.1 KRK00224.1 KQ971311 EEZ98879.1 KQ435803 KOX73092.1 GFTR01006948 JAW09478.1 GEZM01048925 JAV76277.1 GEZM01048936 JAV76264.1 GEZM01048932 JAV76268.1 GEDC01023752 JAS13546.1 KB631840 ERL86606.1 APGK01049324 KB741149 ENN73680.1 GL888102 EGI67532.1 LJIG01001848 KRT85092.1 KK854307 PTY14440.1 KYN05805.1 KYN05807.1 KQ981606 KYN39670.1 KYB29460.1 KQ978081 KYM97095.1 ADTU01026274 GDHC01011273 GDHC01006315 JAQ07356.1 JAQ12314.1 KYQ46343.1 KQ976425 KYM88055.1 KYN05804.1 KYN40875.1 KYM81654.1 KQ978782 KYN28461.1 ADTU01027162 ADTU01027163 ADTU01027164 KYN39671.1 KYN40874.1 ADTU01017950 ADTU01017951
RSAL01000132 RVE46368.1 KQ977104 KYN05808.1 KQ983238 KYQ46342.1 KZ288266 PBC30248.1 KQ981512 KYN40876.1 KK853097 KDR11427.1 GL888284 EGI63268.1 KQ976532 KYM81655.1 KQ978884 KYN27677.1 ADTU01027161 GL451937 EFN78175.1 NEVH01003746 PNF40181.1 PNF40182.1 AJWK01000310 AJWK01000311 AJWK01000312 AJWK01000313 AJWK01000314 CVRI01000047 CRK98393.1 KQ434948 KZC12393.1 GFDF01009258 JAV04826.1 GFDF01009259 JAV04825.1 GDHF01025120 JAI27194.1 PYGN01000948 PSN39063.1 GDHF01026511 GDHF01003173 JAI25803.1 JAI49141.1 GDHF01022509 JAI29805.1 GAKP01011016 JAC47936.1 GAMC01020117 JAB86438.1 JRES01000264 KNC32853.1 KA645081 AFP59710.1 OUUW01000001 SPP72891.1 CCAG010014016 JXJN01022164 JXJN01022165 CH963846 EDW72578.1 GL766597 EFZ15023.1 CH954179 EDV55797.1 GFDF01009385 JAV04699.1 GBXI01013198 GBXI01012822 GBXI01007590 GBXI01005757 JAD01094.1 JAD01470.1 JAD06702.1 JAD08535.1 CM000362 CM002911 EDX07322.1 KMY94211.1 AE013599 AY121685 AAF58040.2 AAM52012.1 GFDF01009386 JAV04698.1 CH480816 EDW48100.1 CH940648 EDW60785.1 KRF79615.1 KRF79616.1 KRF79617.1 KRF79618.1 CH479183 EDW36205.1 CH902619 EDV35823.1 KPU75843.1 KPU75844.1 KPU75845.1 KPU75846.1 CH916367 EDW00756.1 CM000071 EDY69160.1 KRT02513.1 AAZX01004900 CH933808 EDW10197.1 KRG05084.1 KRG05085.1 KRG05086.1 CP012524 ALC42236.1 CM000158 EDW91966.1 KRK00222.1 KRK00223.1 KRK00224.1 KQ971311 EEZ98879.1 KQ435803 KOX73092.1 GFTR01006948 JAW09478.1 GEZM01048925 JAV76277.1 GEZM01048936 JAV76264.1 GEZM01048932 JAV76268.1 GEDC01023752 JAS13546.1 KB631840 ERL86606.1 APGK01049324 KB741149 ENN73680.1 GL888102 EGI67532.1 LJIG01001848 KRT85092.1 KK854307 PTY14440.1 KYN05805.1 KYN05807.1 KQ981606 KYN39670.1 KYB29460.1 KQ978081 KYM97095.1 ADTU01026274 GDHC01011273 GDHC01006315 JAQ07356.1 JAQ12314.1 KYQ46343.1 KQ976425 KYM88055.1 KYN05804.1 KYN40875.1 KYM81654.1 KQ978782 KYN28461.1 ADTU01027162 ADTU01027163 ADTU01027164 KYN39671.1 KYN40874.1 ADTU01017950 ADTU01017951
Proteomes
UP000005204
UP000053240
UP000283053
UP000078542
UP000075809
UP000005203
+ More
UP000242457 UP000078541 UP000027135 UP000007755 UP000078540 UP000078492 UP000005205 UP000008237 UP000235965 UP000092461 UP000183832 UP000076502 UP000095301 UP000245037 UP000037069 UP000268350 UP000092444 UP000092460 UP000092443 UP000007798 UP000078200 UP000008711 UP000000304 UP000095300 UP000000803 UP000001292 UP000192221 UP000008792 UP000192223 UP000008744 UP000007801 UP000001070 UP000001819 UP000002358 UP000009192 UP000092553 UP000002282 UP000007266 UP000053105 UP000030742 UP000019118
UP000242457 UP000078541 UP000027135 UP000007755 UP000078540 UP000078492 UP000005205 UP000008237 UP000235965 UP000092461 UP000183832 UP000076502 UP000095301 UP000245037 UP000037069 UP000268350 UP000092444 UP000092460 UP000092443 UP000007798 UP000078200 UP000008711 UP000000304 UP000095300 UP000000803 UP000001292 UP000192221 UP000008792 UP000192223 UP000008744 UP000007801 UP000001070 UP000001819 UP000002358 UP000009192 UP000092553 UP000002282 UP000007266 UP000053105 UP000030742 UP000019118
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JRH7
A0A2H1VJP3
A0A0N1I937
A0A3S2PB24
A0A151ILP2
A0A151WEV2
+ More
A0A088A4G5 A0A2A3EEV4 A0A195FKJ5 A0A067QRS4 F4WR41 A0A195BBS0 A0A151JNH3 A0A158NVH6 E2C1J9 A0A2J7RH72 A0A2J7RH69 A0A1B0C8B2 A0A1J1IIW4 A0A154PKN9 A0A1L8DEF4 A0A1L8DEL6 A0A0K8UKI2 A0A1I8MUF3 A0A2P8Y492 A0A0K8UGX1 A0A0K8UTZ9 A0A034VX86 W8APK8 A0A0L0CKC8 T1P8J8 A0A3B0JJV7 A0A1B0GBJ0 A0A1B0BXB7 A0A1A9XEC7 B4MKD9 A0A1A9VE41 E9IWP5 B3NPV0 A0A1L8DDZ6 A0A0A1XC57 B4QHH7 A0A1I8NSP7 Q8MRB2 A0A1L8DE28 B4HST6 A0A1W4VCP7 B4LKY1 A0A1W4WDG2 B4GI85 B3MH64 B4J5J3 B5DZ61 K7J5C1 B4KPS0 A0A0M5IXW4 B4P6B6 D6WCF4 A0A1W4WEF1 A0A0N0BFA9 A0A224XI40 A0A1Y1LZ67 A0A1Y1LRF2 A0A1Y1LTU5 A0A1B6CJ75 U4U7K6 N6TWM6 F4WE94 A0A0T6BCR0 A0A2R7W644 A0A1W4X5L7 A0A1W4X6V3 A0A151ILN9 A0A151ILP7 A0A195FG52 A0A139WN88 A0A195C8R6 A0A158NUB4 A0A146LKJ3 A0A151WES1 A0A151I5A5 A0A151ILR1 A0A195FJN8 A0A195BBT0 A0A195EJZ4 A0A158NVI0 A0A195FH24 A0A195FLK8 A0A158NJJ1
A0A088A4G5 A0A2A3EEV4 A0A195FKJ5 A0A067QRS4 F4WR41 A0A195BBS0 A0A151JNH3 A0A158NVH6 E2C1J9 A0A2J7RH72 A0A2J7RH69 A0A1B0C8B2 A0A1J1IIW4 A0A154PKN9 A0A1L8DEF4 A0A1L8DEL6 A0A0K8UKI2 A0A1I8MUF3 A0A2P8Y492 A0A0K8UGX1 A0A0K8UTZ9 A0A034VX86 W8APK8 A0A0L0CKC8 T1P8J8 A0A3B0JJV7 A0A1B0GBJ0 A0A1B0BXB7 A0A1A9XEC7 B4MKD9 A0A1A9VE41 E9IWP5 B3NPV0 A0A1L8DDZ6 A0A0A1XC57 B4QHH7 A0A1I8NSP7 Q8MRB2 A0A1L8DE28 B4HST6 A0A1W4VCP7 B4LKY1 A0A1W4WDG2 B4GI85 B3MH64 B4J5J3 B5DZ61 K7J5C1 B4KPS0 A0A0M5IXW4 B4P6B6 D6WCF4 A0A1W4WEF1 A0A0N0BFA9 A0A224XI40 A0A1Y1LZ67 A0A1Y1LRF2 A0A1Y1LTU5 A0A1B6CJ75 U4U7K6 N6TWM6 F4WE94 A0A0T6BCR0 A0A2R7W644 A0A1W4X5L7 A0A1W4X6V3 A0A151ILN9 A0A151ILP7 A0A195FG52 A0A139WN88 A0A195C8R6 A0A158NUB4 A0A146LKJ3 A0A151WES1 A0A151I5A5 A0A151ILR1 A0A195FJN8 A0A195BBT0 A0A195EJZ4 A0A158NVI0 A0A195FH24 A0A195FLK8 A0A158NJJ1
Ontologies
KEGG
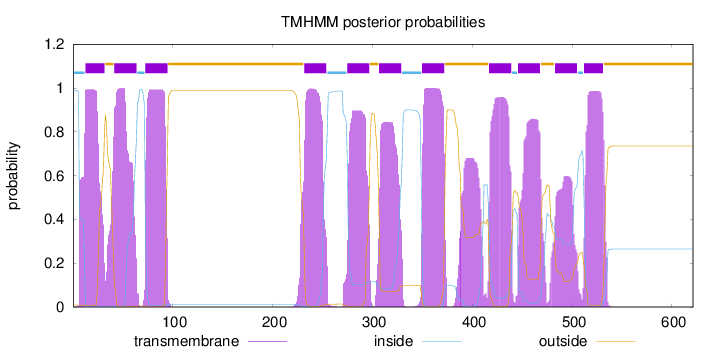
Topology
Length:
621
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
233.63276
Exp number, first 60 AAs:
40.02826
Total prob of N-in:
0.99070
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 41
TMhelix
42 - 64
inside
65 - 72
TMhelix
73 - 95
outside
96 - 231
TMhelix
232 - 254
inside
255 - 274
TMhelix
275 - 297
outside
298 - 306
TMhelix
307 - 329
inside
330 - 349
TMhelix
350 - 372
outside
373 - 416
TMhelix
417 - 439
inside
440 - 445
TMhelix
446 - 468
outside
469 - 482
TMhelix
483 - 505
inside
506 - 511
TMhelix
512 - 531
outside
532 - 621
Population Genetic Test Statistics
Pi
5.369732
Theta
17.389112
Tajima's D
-1.411919
CLR
0.105418
CSRT
0.0671966401679916
Interpretation
Uncertain
