Gene
KWMTBOMO16519
Pre Gene Modal
BGIBMGA012212
Annotation
PREDICTED:_charged_multivesicular_body_protein_7_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.361
Sequence
CDS
ATGCATAGCTCTGCTCTTCTTGGGCTTACGCTAATAGGTATTATAACTCAAACAGCTGAACCAGATAAGAAAGCCACCCCCGTGACAGAGTCAGATGAAGCCCTTGTGAAGCTAGTCACAGCTGAGAGGCACTTGCACGGAGACTCCACAAGACTGTCCCGAGAACTAGCAACTATTGAGAATGATGCTCGAGCTGCACTTCAGCTAGGTAATAGGATTGCGGCCAAGAATCATCTGAGACGTAAGCACAAAGTGCAACGGAAACTGGAACAGTGCACGAAAGCTCTGGAAAACGTACAGAATTTACTACAACAGATGAGGGATGTACATTTCAATGCATCAATTGTCGATACTTACAAGACAACATCAGAAGCTATGAAGCGAGGATTAAAAGACAACGGTCTTGAGGTGGATACAGTTCACGACACGATGGACGATCTGAAAGAGGTAATGGATTCATATAGCGAAATGGAGAGAGCACTTGGTACAACAATAGACGACACCGACACAGCTGAGCTTGAAAGAGAATTGAAGGAACTGCTGTCGGGACCGTCGGCGCCTGGTGGCGGAACTCCCGGGAAGGAGGGGGAGGTGAAGGAATAA
Protein
MHSSALLGLTLIGIITQTAEPDKKATPVTESDEALVKLVTAERHLHGDSTRLSRELATIENDARAALQLGNRIAAKNHLRRKHKVQRKLEQCTKALENVQNLLQQMRDVHFNASIVDTYKTTSEAMKRGLKDNGLEVDTVHDTMDDLKEVMDSYSEMERALGTTIDDTDTAELERELKELLSGPSAPGGGTPGKEGEVKE
Summary
Uniprot
A0A2A4JL95
A0A3S2NFQ0
A0A2W1BJP7
B0WFW4
A0A2S2NB11
J9JTF6
+ More
A0A1Q3F7D4 A0A154PEC2 A0A232EKP0 K7IPF6 A0A084W184 U4UUL4 A0A182GRB1 V5H4X7 A0A1J1J023 A0A1S3D855 A0A088AS34 N6U305 J3JWH1 Q16F51 Q16P72 A0A182RJU3 A0A182WAY9 A0A2M4AJG2 A0A2L2Y6A8 A0A2M4CIQ5 A0A182PVB5 A0A1W5B7G0 A0A195EZ66 A0A2M4CJZ3 A0A195CFJ8 A0A2A3E849 A0A1B6F3N6 A0A2S2QMR1 A0A182NQB8 A0A336MSN9 A0A1S3IJS9 A0A1B6CL40 A0A151J6B3 A0A158NYV4 F4W9T3 A0A310S7L8 T1J0Y3 A0A182SNA2 A0A0K0DIC0 A0A151WKH0 D6WCG1 A0A0L7RG44 A0A195BS09 A0A2J7RPS3 A0A2H2IIF6 A0A026WCP7 A0A0M9A5D6 A0A2J7RPT6 A0A182FT74 E2A5S8 F6ZFT6 A0A2J7RPT7 A0A0A1WZX0 C3Y5U4 A0A182INU5 A0A336L5J9 A0A2G8L8I7 Q22718 A0A0J7L3K2 A0A182MBU3 A0A2M3ZIT0 A0A2M3ZIS7 A0A0V0GAA9 E0VG15 A0A1I7UAZ8 A0A2P6KX35 A0A0C2GRJ9 A0A1I8NXI1 A0A182QIJ9 W4Y2H9 A0A3B0KTV5 W8C2U3 A0A023F7L9 A0A016UQA1 A0A2M4BQC7 A0A016UP83 A8WSG3 G0NP07 K7GIA6 A0A2G5V177 A0A0B1SG72 B3NIK7 G3VRQ4
A0A1Q3F7D4 A0A154PEC2 A0A232EKP0 K7IPF6 A0A084W184 U4UUL4 A0A182GRB1 V5H4X7 A0A1J1J023 A0A1S3D855 A0A088AS34 N6U305 J3JWH1 Q16F51 Q16P72 A0A182RJU3 A0A182WAY9 A0A2M4AJG2 A0A2L2Y6A8 A0A2M4CIQ5 A0A182PVB5 A0A1W5B7G0 A0A195EZ66 A0A2M4CJZ3 A0A195CFJ8 A0A2A3E849 A0A1B6F3N6 A0A2S2QMR1 A0A182NQB8 A0A336MSN9 A0A1S3IJS9 A0A1B6CL40 A0A151J6B3 A0A158NYV4 F4W9T3 A0A310S7L8 T1J0Y3 A0A182SNA2 A0A0K0DIC0 A0A151WKH0 D6WCG1 A0A0L7RG44 A0A195BS09 A0A2J7RPS3 A0A2H2IIF6 A0A026WCP7 A0A0M9A5D6 A0A2J7RPT6 A0A182FT74 E2A5S8 F6ZFT6 A0A2J7RPT7 A0A0A1WZX0 C3Y5U4 A0A182INU5 A0A336L5J9 A0A2G8L8I7 Q22718 A0A0J7L3K2 A0A182MBU3 A0A2M3ZIT0 A0A2M3ZIS7 A0A0V0GAA9 E0VG15 A0A1I7UAZ8 A0A2P6KX35 A0A0C2GRJ9 A0A1I8NXI1 A0A182QIJ9 W4Y2H9 A0A3B0KTV5 W8C2U3 A0A023F7L9 A0A016UQA1 A0A2M4BQC7 A0A016UP83 A8WSG3 G0NP07 K7GIA6 A0A2G5V177 A0A0B1SG72 B3NIK7 G3VRQ4
Pubmed
EMBL
NWSH01001067
PCG72827.1
RSAL01000132
RVE46375.1
KZ150102
PZC73487.1
+ More
DS231921 EDS26499.1 GGMR01001775 MBY14394.1 ABLF02032620 GFDL01011569 JAV23476.1 KQ434887 KZC10212.1 NNAY01003732 OXU18930.1 ATLV01019246 KE525266 KFB43978.1 KB632375 ERL93876.1 JXUM01016395 KQ560457 KXJ82319.1 GALX01000603 JAB67863.1 CVRI01000063 CRL04766.1 APGK01051693 APGK01051694 KB741201 ENN72952.1 BT127589 AEE62551.1 CH478510 EAT32866.1 CH477792 EAT36162.1 GGFK01007602 MBW40923.1 IAAA01017265 LAA03712.1 GGFL01001039 MBW65217.1 KQ981909 KYN33169.1 GGFL01001040 MBW65218.1 KQ977876 KYM99091.1 KZ288329 PBC27927.1 GECZ01024873 JAS44896.1 GGMS01009229 MBY78432.1 UFQT01001639 SSX31287.1 GEDC01023114 GEDC01019449 JAS14184.1 JAS17849.1 KQ979868 KYN18745.1 ADTU01000801 GL888033 EGI69069.1 KQ765323 KQ759877 OAD53989.1 OAD62272.1 JH431742 KQ983012 KYQ48336.1 KQ971311 EEZ99020.1 KQ414599 KOC69794.1 KQ976423 KYM88792.1 NEVH01001355 PNF42842.1 KK107315 QOIP01000002 EZA52824.1 RLU25894.1 KQ435750 KOX76283.1 PNF42843.1 GL437059 EFN71200.1 EAAA01002610 PNF42844.1 GBXI01010354 JAD03938.1 GG666487 EEN64341.1 UFQS01002092 UFQT01002092 SSX13206.1 SSX32645.1 MRZV01000170 PIK56578.1 BX284602 CAA92759.1 LBMM01000802 KMQ97457.1 AXCM01004171 GGFM01007671 MBW28422.1 GGFM01007680 MBW28431.1 GECL01001131 JAP04993.1 DS235131 EEB12321.1 MWRG01004044 PRD30890.1 KN728360 KIH63910.1 AXCN02001893 AAGJ04077408 OUUW01000012 SPP87338.1 GAMC01006389 JAC00167.1 GBBI01001513 JAC17199.1 KE124792 JARK01001369 EPB79523.1 EYC16673.1 GGFJ01006032 MBW55173.1 EYC16672.1 HE601438 CAP23422.2 GL379918 EGT35085.1 AGCU01168837 AGCU01168838 PDUG01000002 PIC45510.1 KN581729 KHJ82160.1 CH954178 EDV52503.1 AEFK01028569 AEFK01028570
DS231921 EDS26499.1 GGMR01001775 MBY14394.1 ABLF02032620 GFDL01011569 JAV23476.1 KQ434887 KZC10212.1 NNAY01003732 OXU18930.1 ATLV01019246 KE525266 KFB43978.1 KB632375 ERL93876.1 JXUM01016395 KQ560457 KXJ82319.1 GALX01000603 JAB67863.1 CVRI01000063 CRL04766.1 APGK01051693 APGK01051694 KB741201 ENN72952.1 BT127589 AEE62551.1 CH478510 EAT32866.1 CH477792 EAT36162.1 GGFK01007602 MBW40923.1 IAAA01017265 LAA03712.1 GGFL01001039 MBW65217.1 KQ981909 KYN33169.1 GGFL01001040 MBW65218.1 KQ977876 KYM99091.1 KZ288329 PBC27927.1 GECZ01024873 JAS44896.1 GGMS01009229 MBY78432.1 UFQT01001639 SSX31287.1 GEDC01023114 GEDC01019449 JAS14184.1 JAS17849.1 KQ979868 KYN18745.1 ADTU01000801 GL888033 EGI69069.1 KQ765323 KQ759877 OAD53989.1 OAD62272.1 JH431742 KQ983012 KYQ48336.1 KQ971311 EEZ99020.1 KQ414599 KOC69794.1 KQ976423 KYM88792.1 NEVH01001355 PNF42842.1 KK107315 QOIP01000002 EZA52824.1 RLU25894.1 KQ435750 KOX76283.1 PNF42843.1 GL437059 EFN71200.1 EAAA01002610 PNF42844.1 GBXI01010354 JAD03938.1 GG666487 EEN64341.1 UFQS01002092 UFQT01002092 SSX13206.1 SSX32645.1 MRZV01000170 PIK56578.1 BX284602 CAA92759.1 LBMM01000802 KMQ97457.1 AXCM01004171 GGFM01007671 MBW28422.1 GGFM01007680 MBW28431.1 GECL01001131 JAP04993.1 DS235131 EEB12321.1 MWRG01004044 PRD30890.1 KN728360 KIH63910.1 AXCN02001893 AAGJ04077408 OUUW01000012 SPP87338.1 GAMC01006389 JAC00167.1 GBBI01001513 JAC17199.1 KE124792 JARK01001369 EPB79523.1 EYC16673.1 GGFJ01006032 MBW55173.1 EYC16672.1 HE601438 CAP23422.2 GL379918 EGT35085.1 AGCU01168837 AGCU01168838 PDUG01000002 PIC45510.1 KN581729 KHJ82160.1 CH954178 EDV52503.1 AEFK01028569 AEFK01028570
Proteomes
UP000218220
UP000283053
UP000002320
UP000007819
UP000076502
UP000215335
+ More
UP000002358 UP000030765 UP000030742 UP000069940 UP000249989 UP000183832 UP000079169 UP000005203 UP000019118 UP000008820 UP000075900 UP000075920 UP000075885 UP000078541 UP000078542 UP000242457 UP000075884 UP000085678 UP000078492 UP000005205 UP000007755 UP000075901 UP000035642 UP000075809 UP000007266 UP000053825 UP000078540 UP000235965 UP000005237 UP000053097 UP000279307 UP000053105 UP000069272 UP000000311 UP000008144 UP000001554 UP000075880 UP000230750 UP000001940 UP000036403 UP000075883 UP000009046 UP000095282 UP000095300 UP000075886 UP000007110 UP000268350 UP000024635 UP000008549 UP000008068 UP000007267 UP000230233 UP000008711 UP000007648
UP000002358 UP000030765 UP000030742 UP000069940 UP000249989 UP000183832 UP000079169 UP000005203 UP000019118 UP000008820 UP000075900 UP000075920 UP000075885 UP000078541 UP000078542 UP000242457 UP000075884 UP000085678 UP000078492 UP000005205 UP000007755 UP000075901 UP000035642 UP000075809 UP000007266 UP000053825 UP000078540 UP000235965 UP000005237 UP000053097 UP000279307 UP000053105 UP000069272 UP000000311 UP000008144 UP000001554 UP000075880 UP000230750 UP000001940 UP000036403 UP000075883 UP000009046 UP000095282 UP000095300 UP000075886 UP000007110 UP000268350 UP000024635 UP000008549 UP000008068 UP000007267 UP000230233 UP000008711 UP000007648
ProteinModelPortal
A0A2A4JL95
A0A3S2NFQ0
A0A2W1BJP7
B0WFW4
A0A2S2NB11
J9JTF6
+ More
A0A1Q3F7D4 A0A154PEC2 A0A232EKP0 K7IPF6 A0A084W184 U4UUL4 A0A182GRB1 V5H4X7 A0A1J1J023 A0A1S3D855 A0A088AS34 N6U305 J3JWH1 Q16F51 Q16P72 A0A182RJU3 A0A182WAY9 A0A2M4AJG2 A0A2L2Y6A8 A0A2M4CIQ5 A0A182PVB5 A0A1W5B7G0 A0A195EZ66 A0A2M4CJZ3 A0A195CFJ8 A0A2A3E849 A0A1B6F3N6 A0A2S2QMR1 A0A182NQB8 A0A336MSN9 A0A1S3IJS9 A0A1B6CL40 A0A151J6B3 A0A158NYV4 F4W9T3 A0A310S7L8 T1J0Y3 A0A182SNA2 A0A0K0DIC0 A0A151WKH0 D6WCG1 A0A0L7RG44 A0A195BS09 A0A2J7RPS3 A0A2H2IIF6 A0A026WCP7 A0A0M9A5D6 A0A2J7RPT6 A0A182FT74 E2A5S8 F6ZFT6 A0A2J7RPT7 A0A0A1WZX0 C3Y5U4 A0A182INU5 A0A336L5J9 A0A2G8L8I7 Q22718 A0A0J7L3K2 A0A182MBU3 A0A2M3ZIT0 A0A2M3ZIS7 A0A0V0GAA9 E0VG15 A0A1I7UAZ8 A0A2P6KX35 A0A0C2GRJ9 A0A1I8NXI1 A0A182QIJ9 W4Y2H9 A0A3B0KTV5 W8C2U3 A0A023F7L9 A0A016UQA1 A0A2M4BQC7 A0A016UP83 A8WSG3 G0NP07 K7GIA6 A0A2G5V177 A0A0B1SG72 B3NIK7 G3VRQ4
A0A1Q3F7D4 A0A154PEC2 A0A232EKP0 K7IPF6 A0A084W184 U4UUL4 A0A182GRB1 V5H4X7 A0A1J1J023 A0A1S3D855 A0A088AS34 N6U305 J3JWH1 Q16F51 Q16P72 A0A182RJU3 A0A182WAY9 A0A2M4AJG2 A0A2L2Y6A8 A0A2M4CIQ5 A0A182PVB5 A0A1W5B7G0 A0A195EZ66 A0A2M4CJZ3 A0A195CFJ8 A0A2A3E849 A0A1B6F3N6 A0A2S2QMR1 A0A182NQB8 A0A336MSN9 A0A1S3IJS9 A0A1B6CL40 A0A151J6B3 A0A158NYV4 F4W9T3 A0A310S7L8 T1J0Y3 A0A182SNA2 A0A0K0DIC0 A0A151WKH0 D6WCG1 A0A0L7RG44 A0A195BS09 A0A2J7RPS3 A0A2H2IIF6 A0A026WCP7 A0A0M9A5D6 A0A2J7RPT6 A0A182FT74 E2A5S8 F6ZFT6 A0A2J7RPT7 A0A0A1WZX0 C3Y5U4 A0A182INU5 A0A336L5J9 A0A2G8L8I7 Q22718 A0A0J7L3K2 A0A182MBU3 A0A2M3ZIT0 A0A2M3ZIS7 A0A0V0GAA9 E0VG15 A0A1I7UAZ8 A0A2P6KX35 A0A0C2GRJ9 A0A1I8NXI1 A0A182QIJ9 W4Y2H9 A0A3B0KTV5 W8C2U3 A0A023F7L9 A0A016UQA1 A0A2M4BQC7 A0A016UP83 A8WSG3 G0NP07 K7GIA6 A0A2G5V177 A0A0B1SG72 B3NIK7 G3VRQ4
PDB
5FD7
E-value=6.90999e-05,
Score=106
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.854722
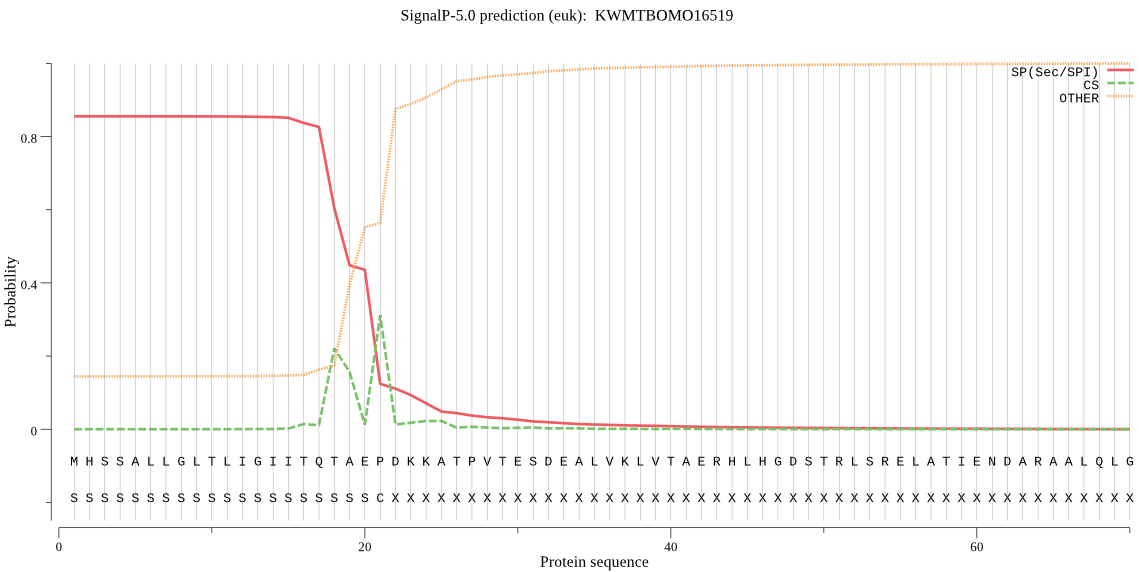
Length:
200
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01765
Exp number, first 60 AAs:
0.01765
Total prob of N-in:
0.01733
outside
1 - 200
Population Genetic Test Statistics
Pi
3.074427
Theta
18.900453
Tajima's D
-1.137184
CLR
0.058963
CSRT
0.110244487775611
Interpretation
Uncertain
