Gene
KWMTBOMO16518 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012213
Annotation
cuticular_protein_RR-1_motif_51_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.425 Extracellular Reliability : 1.016 Nuclear Reliability : 1.587
Sequence
CDS
ATGGCAGTGGCGTCCTTGGGTCAATGTCAACGGCACAGCCAGATAATTCGAGACGACATTGACGGTCCGAACCCGGATGGCTCTTACAAATGGGCAATCGAAACTGATGAGGGAATCTACCACGAGCAGCGAGGGTCGGTGCAAGAAGACACCGGATTGGCTGTGAAGGGGCAGTATCAATACGTCGCTCCCGACGGGCAGGTGATCAACGTACTGTACAGTGCTGACGAGAATGGGTTTCAAGCGAGCGGCGCTCACCTGCCGACACCCCCACCAGTCCCGCCGGCCATACAAAAAATCATTGACTACCTCAACAGCCATCCGACAGAGCCGACTCGGATATAA
Protein
MAVASLGQCQRHSQIIRDDIDGPNPDGSYKWAIETDEGIYHEQRGSVQEDTGLAVKGQYQYVAPDGQVINVLYSADENGFQASGAHLPTPPPVPPAIQKIIDYLNSHPTEPTRI
Summary
Uniprot
C0H6P3
H9JRQ3
A0A3S2NB28
A0A2A4JIC3
A0A2H1W9D6
A0A232F457
+ More
K7JD28 K7J4G3 Q9BPR1 D6WNU9 A0A194Q3B6 C0H6M5 D6WNU8 A0A2A4J2S0 A0A2H1VDC4 A0A2A4IU20 A0A212F0I8 A0A0C9RJI7 Q17G30 A0A1B6JBU0 A0A194R101 A0A1B6GUY0 A0A0L7L4J5 A0A0K8WF58 A0A1W4XP64 A0A0L7QW49 A0A0T6BFM1 A0A182QGA7 A0A0T6BD43 A0A182IU01 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A3EJ46 A0A212FBE3 A0A0J7L9L8 E2A8I2 E2B0E6 A0A3L8DWG0 W8AZ16 A0A0J7L5C1 K7J4G2 A0A182TXD1 A0A182RHG2 U4URD4 N6T2S4 A0A088A4U8 A0A0M9AD49 A0A0A1WRK3 A0A026WNM4 A0A1L8E3U5 A0A182KCU6 A8W7B2 A0A182MIF1 A0A3S2NA82 A0A232FDB1 A0A3G2SFE5 A0A2S1ZS73 A0A026WPJ9 E2BGZ7 A0A2R7WPG2 A0A1B6LNG4 A0A2A3EIE5 I4DIL1 A0A3L8DX45 A0A194Q228 D6WJM7 F4X6E7 I4DKM8 A0A0N1PJL0 A0A195D5A8 A0A0J7NZ86 A0A2S1ZS80 A0A2H8TQ63 A0A151X6J9 A0A194PY11 D6WNV0 A0A222NTA5 A0A1W4XP69 A0A232FDH9 K7J4G1 A0A3S2NQ74 A0A212FBD0 R4G2W3 A0A0C9QI21 E9IFU8 E9IFU9 A0A3L8DVA1 A0A026WR46 A0A195DXL6 A0A182HRK7 A0A182WGU5 A0A0P5S8W5 A0A195BCK5 I4DMG1 A0A182NUJ4 A0A2H1V805
K7JD28 K7J4G3 Q9BPR1 D6WNU9 A0A194Q3B6 C0H6M5 D6WNU8 A0A2A4J2S0 A0A2H1VDC4 A0A2A4IU20 A0A212F0I8 A0A0C9RJI7 Q17G30 A0A1B6JBU0 A0A194R101 A0A1B6GUY0 A0A0L7L4J5 A0A0K8WF58 A0A1W4XP64 A0A0L7QW49 A0A0T6BFM1 A0A182QGA7 A0A0T6BD43 A0A182IU01 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A3EJ46 A0A212FBE3 A0A0J7L9L8 E2A8I2 E2B0E6 A0A3L8DWG0 W8AZ16 A0A0J7L5C1 K7J4G2 A0A182TXD1 A0A182RHG2 U4URD4 N6T2S4 A0A088A4U8 A0A0M9AD49 A0A0A1WRK3 A0A026WNM4 A0A1L8E3U5 A0A182KCU6 A8W7B2 A0A182MIF1 A0A3S2NA82 A0A232FDB1 A0A3G2SFE5 A0A2S1ZS73 A0A026WPJ9 E2BGZ7 A0A2R7WPG2 A0A1B6LNG4 A0A2A3EIE5 I4DIL1 A0A3L8DX45 A0A194Q228 D6WJM7 F4X6E7 I4DKM8 A0A0N1PJL0 A0A195D5A8 A0A0J7NZ86 A0A2S1ZS80 A0A2H8TQ63 A0A151X6J9 A0A194PY11 D6WNV0 A0A222NTA5 A0A1W4XP69 A0A232FDH9 K7J4G1 A0A3S2NQ74 A0A212FBD0 R4G2W3 A0A0C9QI21 E9IFU8 E9IFU9 A0A3L8DVA1 A0A026WR46 A0A195DXL6 A0A182HRK7 A0A182WGU5 A0A0P5S8W5 A0A195BCK5 I4DMG1 A0A182NUJ4 A0A2H1V805
Pubmed
EMBL
BR000552
FAA00554.1
BABH01041912
BABH01041913
RSAL01000132
RVE46369.1
+ More
NWSH01001454 PCG71180.1 ODYU01007168 SOQ49709.1 NNAY01001070 OXU25250.1 AB047488 BR000546 BAB32485.1 FAA00548.1 KQ971343 EFA03209.1 KQ459586 KPI97900.1 BABH01013052 BR000534 FAA00536.1 EFA03210.1 NWSH01003794 PCG65814.1 ODYU01001934 SOQ38827.1 NWSH01007872 PCG62774.1 AGBW02011079 OWR47243.1 GBYB01013422 JAG83189.1 CH477267 EAT45440.1 GECU01011062 JAS96644.1 KQ460883 KPJ11382.1 GECZ01020550 GECZ01003527 JAS49219.1 JAS66242.1 JTDY01002969 KOB70397.1 GDHF01033135 GDHF01028069 GDHF01025247 GDHF01010731 GDHF01010472 GDHF01002521 JAI19179.1 JAI24245.1 JAI27067.1 JAI41583.1 JAI41842.1 JAI49793.1 KQ414716 KOC62801.1 LJIG01000961 KRT85979.1 AXCN02002061 LJIG01001715 KRT85225.1 AAAB01008980 EAA14379.5 KZ288251 PBC31051.1 AGBW02009345 OWR51056.1 LBMM01000180 KMR04587.1 GL437616 EFN70252.1 GL444552 EFN60841.1 QOIP01000003 RLU24652.1 GAMC01016447 JAB90108.1 LBMM01000707 KMQ97684.1 KI207368 ERL95667.1 APGK01046791 KB741067 ENN74424.1 KQ435696 KOX80823.1 GBXI01013036 JAD01256.1 KK107144 EZA57573.1 GFDF01000889 JAV13195.1 EU194558 MG601569 ABW74143.1 AYA49894.1 AXCM01002738 RSAL01000156 RVE45630.1 NNAY01000425 OXU28500.1 MG874710 AYO46607.1 MF942789 AWK28312.1 EZA57576.1 GL448255 EFN85030.1 KK855214 PTY21474.1 GEBQ01014923 GEBQ01001739 JAT25054.1 JAT38238.1 PBC31052.1 AK401129 BAM17751.1 RLU24358.1 KQ459564 KPI99622.1 EFA04466.1 GL888812 EGI57987.1 AK401846 BAM18468.1 KQ460400 KPJ14975.1 KQ976818 KYN08085.1 KMQ97685.1 MF942791 AWK28314.1 GFXV01003593 MBW15398.1 KQ982482 KYQ55918.1 KPI97913.1 EFA03208.2 KX503035 ASQ42721.1 OXU28499.1 RVE45634.1 OWR51052.1 ACPB03023185 GAHY01002236 JAA75274.1 GBYB01000207 JAG69974.1 GL762903 EFZ20536.1 EFZ20555.1 RLU24357.1 EZA57574.1 KQ980155 KYN17457.1 APCN01001715 GDIQ01095106 JAL56620.1 KQ976526 KYM81952.1 AK402479 BAM19101.1 ODYU01001157 SOQ36975.1
NWSH01001454 PCG71180.1 ODYU01007168 SOQ49709.1 NNAY01001070 OXU25250.1 AB047488 BR000546 BAB32485.1 FAA00548.1 KQ971343 EFA03209.1 KQ459586 KPI97900.1 BABH01013052 BR000534 FAA00536.1 EFA03210.1 NWSH01003794 PCG65814.1 ODYU01001934 SOQ38827.1 NWSH01007872 PCG62774.1 AGBW02011079 OWR47243.1 GBYB01013422 JAG83189.1 CH477267 EAT45440.1 GECU01011062 JAS96644.1 KQ460883 KPJ11382.1 GECZ01020550 GECZ01003527 JAS49219.1 JAS66242.1 JTDY01002969 KOB70397.1 GDHF01033135 GDHF01028069 GDHF01025247 GDHF01010731 GDHF01010472 GDHF01002521 JAI19179.1 JAI24245.1 JAI27067.1 JAI41583.1 JAI41842.1 JAI49793.1 KQ414716 KOC62801.1 LJIG01000961 KRT85979.1 AXCN02002061 LJIG01001715 KRT85225.1 AAAB01008980 EAA14379.5 KZ288251 PBC31051.1 AGBW02009345 OWR51056.1 LBMM01000180 KMR04587.1 GL437616 EFN70252.1 GL444552 EFN60841.1 QOIP01000003 RLU24652.1 GAMC01016447 JAB90108.1 LBMM01000707 KMQ97684.1 KI207368 ERL95667.1 APGK01046791 KB741067 ENN74424.1 KQ435696 KOX80823.1 GBXI01013036 JAD01256.1 KK107144 EZA57573.1 GFDF01000889 JAV13195.1 EU194558 MG601569 ABW74143.1 AYA49894.1 AXCM01002738 RSAL01000156 RVE45630.1 NNAY01000425 OXU28500.1 MG874710 AYO46607.1 MF942789 AWK28312.1 EZA57576.1 GL448255 EFN85030.1 KK855214 PTY21474.1 GEBQ01014923 GEBQ01001739 JAT25054.1 JAT38238.1 PBC31052.1 AK401129 BAM17751.1 RLU24358.1 KQ459564 KPI99622.1 EFA04466.1 GL888812 EGI57987.1 AK401846 BAM18468.1 KQ460400 KPJ14975.1 KQ976818 KYN08085.1 KMQ97685.1 MF942791 AWK28314.1 GFXV01003593 MBW15398.1 KQ982482 KYQ55918.1 KPI97913.1 EFA03208.2 KX503035 ASQ42721.1 OXU28499.1 RVE45634.1 OWR51052.1 ACPB03023185 GAHY01002236 JAA75274.1 GBYB01000207 JAG69974.1 GL762903 EFZ20536.1 EFZ20555.1 RLU24357.1 EZA57574.1 KQ980155 KYN17457.1 APCN01001715 GDIQ01095106 JAL56620.1 KQ976526 KYM81952.1 AK402479 BAM19101.1 ODYU01001157 SOQ36975.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000215335
UP000002358
UP000007266
+ More
UP000053268 UP000007151 UP000008820 UP000053240 UP000037510 UP000192223 UP000053825 UP000075886 UP000075880 UP000075903 UP000075882 UP000076407 UP000007062 UP000242457 UP000036403 UP000000311 UP000279307 UP000075902 UP000075900 UP000030742 UP000019118 UP000005203 UP000053105 UP000053097 UP000075881 UP000075883 UP000008237 UP000007755 UP000078542 UP000075809 UP000015103 UP000078492 UP000075840 UP000075920 UP000078540 UP000075884
UP000053268 UP000007151 UP000008820 UP000053240 UP000037510 UP000192223 UP000053825 UP000075886 UP000075880 UP000075903 UP000075882 UP000076407 UP000007062 UP000242457 UP000036403 UP000000311 UP000279307 UP000075902 UP000075900 UP000030742 UP000019118 UP000005203 UP000053105 UP000053097 UP000075881 UP000075883 UP000008237 UP000007755 UP000078542 UP000075809 UP000015103 UP000078492 UP000075840 UP000075920 UP000078540 UP000075884
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6P3
H9JRQ3
A0A3S2NB28
A0A2A4JIC3
A0A2H1W9D6
A0A232F457
+ More
K7JD28 K7J4G3 Q9BPR1 D6WNU9 A0A194Q3B6 C0H6M5 D6WNU8 A0A2A4J2S0 A0A2H1VDC4 A0A2A4IU20 A0A212F0I8 A0A0C9RJI7 Q17G30 A0A1B6JBU0 A0A194R101 A0A1B6GUY0 A0A0L7L4J5 A0A0K8WF58 A0A1W4XP64 A0A0L7QW49 A0A0T6BFM1 A0A182QGA7 A0A0T6BD43 A0A182IU01 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A3EJ46 A0A212FBE3 A0A0J7L9L8 E2A8I2 E2B0E6 A0A3L8DWG0 W8AZ16 A0A0J7L5C1 K7J4G2 A0A182TXD1 A0A182RHG2 U4URD4 N6T2S4 A0A088A4U8 A0A0M9AD49 A0A0A1WRK3 A0A026WNM4 A0A1L8E3U5 A0A182KCU6 A8W7B2 A0A182MIF1 A0A3S2NA82 A0A232FDB1 A0A3G2SFE5 A0A2S1ZS73 A0A026WPJ9 E2BGZ7 A0A2R7WPG2 A0A1B6LNG4 A0A2A3EIE5 I4DIL1 A0A3L8DX45 A0A194Q228 D6WJM7 F4X6E7 I4DKM8 A0A0N1PJL0 A0A195D5A8 A0A0J7NZ86 A0A2S1ZS80 A0A2H8TQ63 A0A151X6J9 A0A194PY11 D6WNV0 A0A222NTA5 A0A1W4XP69 A0A232FDH9 K7J4G1 A0A3S2NQ74 A0A212FBD0 R4G2W3 A0A0C9QI21 E9IFU8 E9IFU9 A0A3L8DVA1 A0A026WR46 A0A195DXL6 A0A182HRK7 A0A182WGU5 A0A0P5S8W5 A0A195BCK5 I4DMG1 A0A182NUJ4 A0A2H1V805
K7JD28 K7J4G3 Q9BPR1 D6WNU9 A0A194Q3B6 C0H6M5 D6WNU8 A0A2A4J2S0 A0A2H1VDC4 A0A2A4IU20 A0A212F0I8 A0A0C9RJI7 Q17G30 A0A1B6JBU0 A0A194R101 A0A1B6GUY0 A0A0L7L4J5 A0A0K8WF58 A0A1W4XP64 A0A0L7QW49 A0A0T6BFM1 A0A182QGA7 A0A0T6BD43 A0A182IU01 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A3EJ46 A0A212FBE3 A0A0J7L9L8 E2A8I2 E2B0E6 A0A3L8DWG0 W8AZ16 A0A0J7L5C1 K7J4G2 A0A182TXD1 A0A182RHG2 U4URD4 N6T2S4 A0A088A4U8 A0A0M9AD49 A0A0A1WRK3 A0A026WNM4 A0A1L8E3U5 A0A182KCU6 A8W7B2 A0A182MIF1 A0A3S2NA82 A0A232FDB1 A0A3G2SFE5 A0A2S1ZS73 A0A026WPJ9 E2BGZ7 A0A2R7WPG2 A0A1B6LNG4 A0A2A3EIE5 I4DIL1 A0A3L8DX45 A0A194Q228 D6WJM7 F4X6E7 I4DKM8 A0A0N1PJL0 A0A195D5A8 A0A0J7NZ86 A0A2S1ZS80 A0A2H8TQ63 A0A151X6J9 A0A194PY11 D6WNV0 A0A222NTA5 A0A1W4XP69 A0A232FDH9 K7J4G1 A0A3S2NQ74 A0A212FBD0 R4G2W3 A0A0C9QI21 E9IFU8 E9IFU9 A0A3L8DVA1 A0A026WR46 A0A195DXL6 A0A182HRK7 A0A182WGU5 A0A0P5S8W5 A0A195BCK5 I4DMG1 A0A182NUJ4 A0A2H1V805
Ontologies
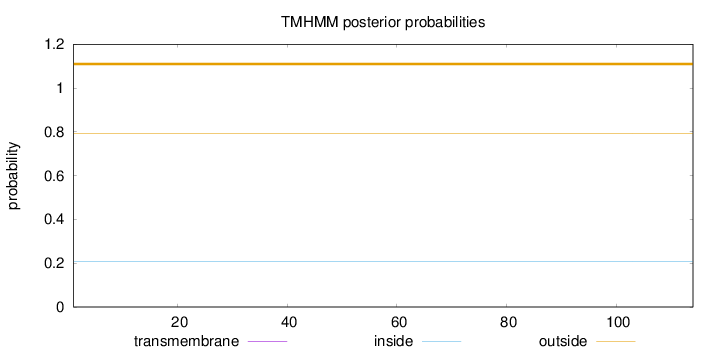
Topology
Length:
114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.20695
outside
1 - 114
Population Genetic Test Statistics
Pi
0.957878
Theta
15.292181
Tajima's D
-2.235661
CLR
30.498763
CSRT
0.00419979001049947
Interpretation
Uncertain
