Gene
KWMTBOMO16509
Pre Gene Modal
BGIBMGA010192
Annotation
Putative_nuclease_HARBI1-like_Protein_[Tribolium_castaneum]
Location in the cell
Nuclear Reliability : 3.074
Sequence
CDS
ATGTCGCTCTCAAGCCTATCGTCGTTGTCTGACCCGGAAGAAGTCGTTTCAACTGGACGTAGAATACCAGATAGGGTAAATATATTTGATGAACATGATGGTGAAGATTTCCGCATTAGATATCGTCTATCTAAAGTTACGGCTCTGCAAATTTCTGAACTTTTGGATTTAGAACCAGTAACACAACGAAATAAAGCCATAGATGGCATAACACAATTATTAATTTGTTTGAGGTTCTTTGCCACTGGATCATTTCAGCAGGTGCTTGGTGATTTAGTTAATATTCATAAATCAACAGTATGTCGTATAGTACAGAAAGTGACACGCAAGCTCGCAGATCTGAGCTTAGCTTATATTAAGATGCCTGATAGGCAGGAGTTGAGAAGGGTTGCAGAAGACTTTTATGAGATTGCTGGATTCCCACGGGTTGCAGGAGCCCTAGATTGCACCCATATCAAAATAGTTAGCCGAGGAGGGTCCTTAGCCGAACTGTATCGTTGCAGAAAAGGATATTTTTCAATTAATGTTCAAGCGGTATGTGATGCCTCTTTAAAAATAAGAGATTTAATTGTAAGGTGGCCAGGATCTGTACATGATTCAACAATATTCAACAACTCCCATTTGTGTGCAGATTTTGAAAGAGGAAAATATAGCCCATACCATTTATTAGGAGATAGTGGCTACTTCAATAAGAACTATTTGCTGACCCCTTTTTTGAATCCTCAAAATTCTGCAGAGGAAGCTTATAATAAAAGCCACATTTCTACTAGAAATACTGTGGAGAGGTGCTTCGGGGTACTTAAACGTCGCTGCCCATGCTTGGAAAAAGGAATAACTTTAAAAAAAACTTCAACCATTCTTCAGGTGATTGTGGCATGTGTAGTGCTACATAACATATGCATAGAGCTAAATGAAGTTGATCCTCCAGAAGACATTGAAATTTCAGTAGACAGTCCTAATATTGGAATATTGTCTATAAGTTCTCAGTCCACAAACAGTGCTGTTCGTGCTGCACTAGTAAATAGTGTGTTTTGTTAA
Protein
MSLSSLSSLSDPEEVVSTGRRIPDRVNIFDEHDGEDFRIRYRLSKVTALQISELLDLEPVTQRNKAIDGITQLLICLRFFATGSFQQVLGDLVNIHKSTVCRIVQKVTRKLADLSLAYIKMPDRQELRRVAEDFYEIAGFPRVAGALDCTHIKIVSRGGSLAELYRCRKGYFSINVQAVCDASLKIRDLIVRWPGSVHDSTIFNNSHLCADFERGKYSPYHLLGDSGYFNKNYLLTPFLNPQNSAEEAYNKSHISTRNTVERCFGVLKRRCPCLEKGITLKKTSTILQVIVACVVLHNICIELNEVDPPEDIEISVDSPNIGILSISSQSTNSAVRAALVNSVFC
Summary
Uniprot
H9JKZ1
D6WYB2
A0A1Y1KFZ4
D7EKQ7
A0A1Y1MC35
J9JTM0
+ More
J9LL64 A0A2P8Z014 J9JM90 A0A3S2P3B6 C4WUD0 N6TYP6 A0A1I8NJZ2 A0A2S2NKK2 N6UD62 A0A0L7LT10 C4WVD2 A0A2S2NS44 J9KWE0 A0A2R2MN13 S4PD89 A0A3S2KYY9 J9M1Z2 J9K8M3 U4U597 A0A1Y1M7D0 J9JXP4 A0A1Y1KQW0 A0A0J7K466 A0A1Y1MZ45 J9KLH0 A0A1S3JPJ3 X1XI09 J9M2S7 J9L523 J9KQN6 J9JZ87 A0A1Y1KLA7 A0A1Y1MBC4 A0A2A4IV58 A0A1Y1MXP7 X1X0S1 J9LYS1 U4U2V2 X1WZQ8 J9LXS6 A0A2S2Q8E2 J9KK24 J9JJW2 A0A0A9ZIH6 J9LUU4 A0A1Y1KRW4 A0A2R5LJM8 N6TRS6 J9L1A7 A0A3Q0J0W5 A0A3S2LMD2 X1XKK1 J9M943 A0A1B6KEC0 J9KAY9 A0A1Y1KVK5 J9L1W5 A0A2P8YDT7 J9KF22 X1WR39 J9LCM8 A0A1E1WP29 J9M4Q3 A0A0L7L248 A0A1E1VZ84 J9LUG4 J9JZI3 A0A1S3J8B0 A0A2P8Y1X7 A0A1E1W4G1 J9LQJ0 A0A158NUI4 A0A147BBZ9 A0A194QSI6 A0A3L8DKW7 A0A0L7L4N7 A0A0A9X6M5 A0A090X8B9 A0A131XTB4 A0A3B1KGA0 A0A2A4IXD7 J9KAE0 J9KPC8 A0A1Y1M620 A0A3B1K7F4 A0A0J7MTD8
J9LL64 A0A2P8Z014 J9JM90 A0A3S2P3B6 C4WUD0 N6TYP6 A0A1I8NJZ2 A0A2S2NKK2 N6UD62 A0A0L7LT10 C4WVD2 A0A2S2NS44 J9KWE0 A0A2R2MN13 S4PD89 A0A3S2KYY9 J9M1Z2 J9K8M3 U4U597 A0A1Y1M7D0 J9JXP4 A0A1Y1KQW0 A0A0J7K466 A0A1Y1MZ45 J9KLH0 A0A1S3JPJ3 X1XI09 J9M2S7 J9L523 J9KQN6 J9JZ87 A0A1Y1KLA7 A0A1Y1MBC4 A0A2A4IV58 A0A1Y1MXP7 X1X0S1 J9LYS1 U4U2V2 X1WZQ8 J9LXS6 A0A2S2Q8E2 J9KK24 J9JJW2 A0A0A9ZIH6 J9LUU4 A0A1Y1KRW4 A0A2R5LJM8 N6TRS6 J9L1A7 A0A3Q0J0W5 A0A3S2LMD2 X1XKK1 J9M943 A0A1B6KEC0 J9KAY9 A0A1Y1KVK5 J9L1W5 A0A2P8YDT7 J9KF22 X1WR39 J9LCM8 A0A1E1WP29 J9M4Q3 A0A0L7L248 A0A1E1VZ84 J9LUG4 J9JZI3 A0A1S3J8B0 A0A2P8Y1X7 A0A1E1W4G1 J9LQJ0 A0A158NUI4 A0A147BBZ9 A0A194QSI6 A0A3L8DKW7 A0A0L7L4N7 A0A0A9X6M5 A0A090X8B9 A0A131XTB4 A0A3B1KGA0 A0A2A4IXD7 J9KAE0 J9KPC8 A0A1Y1M620 A0A3B1K7F4 A0A0J7MTD8
Pubmed
EMBL
BABH01011318
KQ971356
EFA09128.1
GEZM01088054
JAV58326.1
DS497765
+ More
EFA11633.1 GEZM01035464 GEZM01035462 JAV83213.1 ABLF02029655 ABLF02030019 ABLF02019988 ABLF02066411 PYGN01000260 PSN49845.1 ABLF02016431 RSAL01001934 RVE40628.1 ABLF02006541 ABLF02042859 AK341014 BAH71500.1 APGK01005289 KB736163 ENN83248.1 GGMR01005102 MBY17721.1 APGK01040149 KB740975 ENN76582.1 JTDY01000151 KOB78582.1 AK341464 BAH71852.1 GGMR01007355 MBY19974.1 ABLF02016012 ABLF02055237 GAIX01003761 JAA88799.1 RSAL01000416 RVE41817.1 ABLF02028680 ABLF02006612 KB632043 ERL88242.1 GEZM01038551 JAV81614.1 ABLF02008545 GEZM01076717 JAV63704.1 LBMM01014280 KMQ85248.1 GEZM01017851 GEZM01017841 GEZM01017840 JAV90428.1 ABLF02022234 ABLF02058482 ABLF02012281 ABLF02058185 ABLF02011053 ABLF02035722 ABLF02034876 ABLF02036739 GEZM01080383 JAV62179.1 GEZM01038550 JAV81615.1 NWSH01007141 PCG63072.1 GEZM01017837 GEZM01017836 GEZM01017835 JAV90434.1 ABLF02020738 ABLF02012895 KB631844 ERL86663.1 ABLF02008641 ABLF02011808 ABLF02003846 GGMS01004279 MBY73482.1 ABLF02038239 ABLF02011428 GBHO01000839 JAG42765.1 ABLF02007356 GEZM01078171 GEZM01078169 GEZM01078168 GEZM01078162 GEZM01078161 GEZM01078158 GEZM01078157 GEZM01078155 GEZM01078153 GEZM01078152 GEZM01078149 GEZM01078144 GEZM01078140 GEZM01078137 JAV62920.1 GGLE01005627 MBY09753.1 APGK01006027 KB736544 ENN83189.1 ABLF02008686 RSAL01000057 RVE49808.1 ABLF02020736 ABLF02017542 GEBQ01030196 JAT09781.1 ABLF02015906 GEZM01078173 GEZM01078172 GEZM01078170 GEZM01078167 GEZM01078166 GEZM01078160 GEZM01078150 GEZM01078147 GEZM01078145 GEZM01078139 GEZM01078138 GEZM01078136 GEZM01078135 GEZM01078134 JAV62907.1 ABLF02019815 ABLF02019816 PYGN01000674 PSN42418.1 ABLF02061340 ABLF02063305 ABLF02010299 GDQN01002346 JAT88708.1 ABLF02016385 JTDY01003471 KOB69505.1 GDQN01011014 GDQN01002039 JAT80040.1 JAT89015.1 ABLF02024993 ABLF02024998 ABLF02025000 ABLF02007265 PYGN01001035 PSN38265.1 GDQN01011578 GDQN01009283 JAT79476.1 JAT81771.1 ABLF02010163 ADTU01026491 GEGO01007104 JAR88300.1 KQ461175 KPJ07945.1 QOIP01000007 RLU20812.1 JTDY01002978 KOB70380.1 GBHO01028298 JAG15306.1 GBIH01002049 JAC92661.1 GEFM01006224 JAP69572.1 NWSH01005652 PCG64078.1 ABLF02014659 ABLF02034348 GEZM01039795 GEZM01039794 GEZM01039793 GEZM01039792 GEZM01039791 GEZM01039790 GEZM01039789 JAV81183.1 LBMM01018514 KMQ83765.1
EFA11633.1 GEZM01035464 GEZM01035462 JAV83213.1 ABLF02029655 ABLF02030019 ABLF02019988 ABLF02066411 PYGN01000260 PSN49845.1 ABLF02016431 RSAL01001934 RVE40628.1 ABLF02006541 ABLF02042859 AK341014 BAH71500.1 APGK01005289 KB736163 ENN83248.1 GGMR01005102 MBY17721.1 APGK01040149 KB740975 ENN76582.1 JTDY01000151 KOB78582.1 AK341464 BAH71852.1 GGMR01007355 MBY19974.1 ABLF02016012 ABLF02055237 GAIX01003761 JAA88799.1 RSAL01000416 RVE41817.1 ABLF02028680 ABLF02006612 KB632043 ERL88242.1 GEZM01038551 JAV81614.1 ABLF02008545 GEZM01076717 JAV63704.1 LBMM01014280 KMQ85248.1 GEZM01017851 GEZM01017841 GEZM01017840 JAV90428.1 ABLF02022234 ABLF02058482 ABLF02012281 ABLF02058185 ABLF02011053 ABLF02035722 ABLF02034876 ABLF02036739 GEZM01080383 JAV62179.1 GEZM01038550 JAV81615.1 NWSH01007141 PCG63072.1 GEZM01017837 GEZM01017836 GEZM01017835 JAV90434.1 ABLF02020738 ABLF02012895 KB631844 ERL86663.1 ABLF02008641 ABLF02011808 ABLF02003846 GGMS01004279 MBY73482.1 ABLF02038239 ABLF02011428 GBHO01000839 JAG42765.1 ABLF02007356 GEZM01078171 GEZM01078169 GEZM01078168 GEZM01078162 GEZM01078161 GEZM01078158 GEZM01078157 GEZM01078155 GEZM01078153 GEZM01078152 GEZM01078149 GEZM01078144 GEZM01078140 GEZM01078137 JAV62920.1 GGLE01005627 MBY09753.1 APGK01006027 KB736544 ENN83189.1 ABLF02008686 RSAL01000057 RVE49808.1 ABLF02020736 ABLF02017542 GEBQ01030196 JAT09781.1 ABLF02015906 GEZM01078173 GEZM01078172 GEZM01078170 GEZM01078167 GEZM01078166 GEZM01078160 GEZM01078150 GEZM01078147 GEZM01078145 GEZM01078139 GEZM01078138 GEZM01078136 GEZM01078135 GEZM01078134 JAV62907.1 ABLF02019815 ABLF02019816 PYGN01000674 PSN42418.1 ABLF02061340 ABLF02063305 ABLF02010299 GDQN01002346 JAT88708.1 ABLF02016385 JTDY01003471 KOB69505.1 GDQN01011014 GDQN01002039 JAT80040.1 JAT89015.1 ABLF02024993 ABLF02024998 ABLF02025000 ABLF02007265 PYGN01001035 PSN38265.1 GDQN01011578 GDQN01009283 JAT79476.1 JAT81771.1 ABLF02010163 ADTU01026491 GEGO01007104 JAR88300.1 KQ461175 KPJ07945.1 QOIP01000007 RLU20812.1 JTDY01002978 KOB70380.1 GBHO01028298 JAG15306.1 GBIH01002049 JAC92661.1 GEFM01006224 JAP69572.1 NWSH01005652 PCG64078.1 ABLF02014659 ABLF02034348 GEZM01039795 GEZM01039794 GEZM01039793 GEZM01039792 GEZM01039791 GEZM01039790 GEZM01039789 JAV81183.1 LBMM01018514 KMQ83765.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JKZ1
D6WYB2
A0A1Y1KFZ4
D7EKQ7
A0A1Y1MC35
J9JTM0
+ More
J9LL64 A0A2P8Z014 J9JM90 A0A3S2P3B6 C4WUD0 N6TYP6 A0A1I8NJZ2 A0A2S2NKK2 N6UD62 A0A0L7LT10 C4WVD2 A0A2S2NS44 J9KWE0 A0A2R2MN13 S4PD89 A0A3S2KYY9 J9M1Z2 J9K8M3 U4U597 A0A1Y1M7D0 J9JXP4 A0A1Y1KQW0 A0A0J7K466 A0A1Y1MZ45 J9KLH0 A0A1S3JPJ3 X1XI09 J9M2S7 J9L523 J9KQN6 J9JZ87 A0A1Y1KLA7 A0A1Y1MBC4 A0A2A4IV58 A0A1Y1MXP7 X1X0S1 J9LYS1 U4U2V2 X1WZQ8 J9LXS6 A0A2S2Q8E2 J9KK24 J9JJW2 A0A0A9ZIH6 J9LUU4 A0A1Y1KRW4 A0A2R5LJM8 N6TRS6 J9L1A7 A0A3Q0J0W5 A0A3S2LMD2 X1XKK1 J9M943 A0A1B6KEC0 J9KAY9 A0A1Y1KVK5 J9L1W5 A0A2P8YDT7 J9KF22 X1WR39 J9LCM8 A0A1E1WP29 J9M4Q3 A0A0L7L248 A0A1E1VZ84 J9LUG4 J9JZI3 A0A1S3J8B0 A0A2P8Y1X7 A0A1E1W4G1 J9LQJ0 A0A158NUI4 A0A147BBZ9 A0A194QSI6 A0A3L8DKW7 A0A0L7L4N7 A0A0A9X6M5 A0A090X8B9 A0A131XTB4 A0A3B1KGA0 A0A2A4IXD7 J9KAE0 J9KPC8 A0A1Y1M620 A0A3B1K7F4 A0A0J7MTD8
J9LL64 A0A2P8Z014 J9JM90 A0A3S2P3B6 C4WUD0 N6TYP6 A0A1I8NJZ2 A0A2S2NKK2 N6UD62 A0A0L7LT10 C4WVD2 A0A2S2NS44 J9KWE0 A0A2R2MN13 S4PD89 A0A3S2KYY9 J9M1Z2 J9K8M3 U4U597 A0A1Y1M7D0 J9JXP4 A0A1Y1KQW0 A0A0J7K466 A0A1Y1MZ45 J9KLH0 A0A1S3JPJ3 X1XI09 J9M2S7 J9L523 J9KQN6 J9JZ87 A0A1Y1KLA7 A0A1Y1MBC4 A0A2A4IV58 A0A1Y1MXP7 X1X0S1 J9LYS1 U4U2V2 X1WZQ8 J9LXS6 A0A2S2Q8E2 J9KK24 J9JJW2 A0A0A9ZIH6 J9LUU4 A0A1Y1KRW4 A0A2R5LJM8 N6TRS6 J9L1A7 A0A3Q0J0W5 A0A3S2LMD2 X1XKK1 J9M943 A0A1B6KEC0 J9KAY9 A0A1Y1KVK5 J9L1W5 A0A2P8YDT7 J9KF22 X1WR39 J9LCM8 A0A1E1WP29 J9M4Q3 A0A0L7L248 A0A1E1VZ84 J9LUG4 J9JZI3 A0A1S3J8B0 A0A2P8Y1X7 A0A1E1W4G1 J9LQJ0 A0A158NUI4 A0A147BBZ9 A0A194QSI6 A0A3L8DKW7 A0A0L7L4N7 A0A0A9X6M5 A0A090X8B9 A0A131XTB4 A0A3B1KGA0 A0A2A4IXD7 J9KAE0 J9KPC8 A0A1Y1M620 A0A3B1K7F4 A0A0J7MTD8
Ontologies
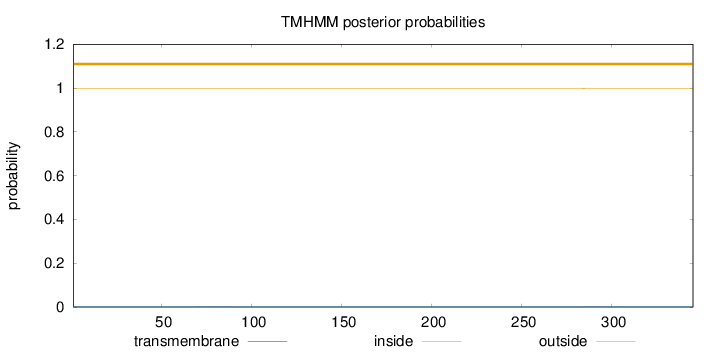
Topology
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02225
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00204
outside
1 - 345
Population Genetic Test Statistics
Pi
13.734212
Theta
21.946903
Tajima's D
-1.152558
CLR
0
CSRT
0.108894555272236
Interpretation
Uncertain
