Gene
KWMTBOMO16500
Pre Gene Modal
BGIBMGA004565
Annotation
PREDICTED:_uncharacterized_protein_LOC101743387_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.125
Sequence
CDS
ATGTCCTTAGCCGAAGGAGCTTCAGACACAGAACAGGCCAGGCAAGAGGAAGATAAGAGCTCGACCACGCTCCAGAAAACCAACGAGAAACGTCAGATGTCCGACTCGTACATGTGGAAAGAACAGATTCACACTCTAATGAAGGGCGGAGAGAGCGAGTCCGACATATCGGACTCGGAGCATTTCCTCACAAAGGGCGCCTTCCTTCTCACTCCCTCCCACGGCCTCAGCATGGAAAAAGCCTCCACCATCTGTGAGAAGATGGAGTTCAAGGGATGCTTCTCCCTGACGAAGACTGCTACTGGAATACTCTTTAAGTTTTCCAGCGTCGATGACTATCAGATGGTATTCAAGAAAGGTTTCCATAAAGTTACCGGATCGCGATTCTATAGGAAGATCGCTATTCCGTGCAGACCGCAGAAGACCTTTACCATTTACGTATACGACATTCCCGATGAGATCCCAGAGGAGGATGTGCGCCACGCTTTGTACAAATTTACTTCAGTCGTTGAGGTCGTCAGGCTGTATTACTCTGGATCCCCAAAAGACGGGAACACTGGTACATCGACCCCAAAACCAGAGAAGACCTCGTCCCGAGCTCTCACGACAATTGATGGAGAGCTCGTGAGCAGCATGGTGCCGAAGGAAGCCACGAGCGTGGTCCGTGTCACTCTCGCCAACATCGAGGAAGCCAACATTCTGCTGCAAAACGGCCTGGATTTCTACGGAGCTACGTTCTTTCCAACCGAATTAGCTACCCCAACCCAGGCAGCTAAGTTGATTAAGCCTAGCAAAGTAACTGGAGGTACCGTGTCAGCCAGAGTCCGTGATTTGCTCCCGGTCTTCGACCAGCAAGGGTTCGCCAAGTTCGCTCCACCGGCCTCTAGACTGGTGAAGCCACGTTCCAAGTGA
Protein
MSLAEGASDTEQARQEEDKSSTTLQKTNEKRQMSDSYMWKEQIHTLMKGGESESDISDSEHFLTKGAFLLTPSHGLSMEKASTICEKMEFKGCFSLTKTATGILFKFSSVDDYQMVFKKGFHKVTGSRFYRKIAIPCRPQKTFTIYVYDIPDEIPEEDVRHALYKFTSVVEVVRLYYSGSPKDGNTGTSTPKPEKTSSRALTTIDGELVSSMVPKEATSVVRVTLANIEEANILLQNGLDFYGATFFPTELATPTQAAKLIKPSKVTGGTVSARVRDLLPVFDQQGFAKFAPPASRLVKPRSK
Summary
Uniprot
A0A2A4JV89
A0A2A4JVM7
A0A2W1BJJ6
H9J4X5
A0A3S2LCE5
A0A194QPX6
+ More
A0A212FCB2 A0A2H1WFL9 I4DK58 A0A0L7KMS4 A0A0L7K3E7 W8CBR0 A0A1J1IRE9 B4L673 A0A034V2A9 A0A0A1X766 A0A0K8VCF4 A0A1I8NF41 A0A0Q9XSK6 A0A0L0CDX7 A0A1B0FGI8 A0A0P9CFF5 A0A2P8ZFZ1 A0A182IR26 A0A1B0BJW1 A0A1A9YPL0 A0A0Q9X8G7 A7US28 A0A084WF90 A0A1A9VC71 A0A1B0A9W1 A0A1L8DD19 A0A2J7RRF9 A0A182USJ1 A0A182XNG1 A0A182IDQ9 A0A1A9WFW4 A0A182TGG9 A0A1B0CG90 A0A0M4F8T1 B3MQ59 A0A067QTL7 A0A1I8NZD9 B4NPD4 A8JUZ6 A0A0P9A5P1 A0A182PKY6 A0A0R1E9B2 A0A182K5G6 A0A0Q5T5B7 E2C7B8 A0A182QY71 A0A182VV53 A0A1W4VG14 A0A182NC44 A0A088AEH6 A0A0R3P1G5 A0A3B0KDJ3 A0A0L7QMW3 B4MAJ2 A0A195FSY9 F4WBZ1 A0A195BKY5 A0A158NZ15 F5HJ22 A0A3L8DWQ9 A0A1W4VSP0 A0A151WV06 A0A1B6LPR9 A0A0M9A0P0 A0A0C9RUC8 A0A3Q0JCK5 Q9W426 A0A182FMH7 A0A0R3P176 A0A3B0KMI6 A0A3B0KR09 A0A0R3P1A0 Q29GF9 A0A0C9RX14 A0A2S2QPM4 A0A2S2PSF8 A0A1W4VFQ8 A0A0R1EFH2 A0A0Q5T5B1 A0A0R3P173 A0A182RM50 A0A3B0KIR2 A0A1W4VTC4 A0A1B6D4C6 A0A023EQN0 A0A2A3EKB2 A0A1S4G7S3 J9KLL3 A0A182Y4T3 E2AAJ6 A0A1W4VU16 B4I0D8 X2JCR6
A0A212FCB2 A0A2H1WFL9 I4DK58 A0A0L7KMS4 A0A0L7K3E7 W8CBR0 A0A1J1IRE9 B4L673 A0A034V2A9 A0A0A1X766 A0A0K8VCF4 A0A1I8NF41 A0A0Q9XSK6 A0A0L0CDX7 A0A1B0FGI8 A0A0P9CFF5 A0A2P8ZFZ1 A0A182IR26 A0A1B0BJW1 A0A1A9YPL0 A0A0Q9X8G7 A7US28 A0A084WF90 A0A1A9VC71 A0A1B0A9W1 A0A1L8DD19 A0A2J7RRF9 A0A182USJ1 A0A182XNG1 A0A182IDQ9 A0A1A9WFW4 A0A182TGG9 A0A1B0CG90 A0A0M4F8T1 B3MQ59 A0A067QTL7 A0A1I8NZD9 B4NPD4 A8JUZ6 A0A0P9A5P1 A0A182PKY6 A0A0R1E9B2 A0A182K5G6 A0A0Q5T5B7 E2C7B8 A0A182QY71 A0A182VV53 A0A1W4VG14 A0A182NC44 A0A088AEH6 A0A0R3P1G5 A0A3B0KDJ3 A0A0L7QMW3 B4MAJ2 A0A195FSY9 F4WBZ1 A0A195BKY5 A0A158NZ15 F5HJ22 A0A3L8DWQ9 A0A1W4VSP0 A0A151WV06 A0A1B6LPR9 A0A0M9A0P0 A0A0C9RUC8 A0A3Q0JCK5 Q9W426 A0A182FMH7 A0A0R3P176 A0A3B0KMI6 A0A3B0KR09 A0A0R3P1A0 Q29GF9 A0A0C9RX14 A0A2S2QPM4 A0A2S2PSF8 A0A1W4VFQ8 A0A0R1EFH2 A0A0Q5T5B1 A0A0R3P173 A0A182RM50 A0A3B0KIR2 A0A1W4VTC4 A0A1B6D4C6 A0A023EQN0 A0A2A3EKB2 A0A1S4G7S3 J9KLL3 A0A182Y4T3 E2AAJ6 A0A1W4VU16 B4I0D8 X2JCR6
Pubmed
28756777
19121390
26354079
22118469
22651552
26227816
+ More
24495485 17994087 25348373 25830018 25315136 26108605 29403074 12364791 24438588 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 15632085 21719571 21347285 30249741 24945155 26483478 17510324 25244985
24495485 17994087 25348373 25830018 25315136 26108605 29403074 12364791 24438588 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 15632085 21719571 21347285 30249741 24945155 26483478 17510324 25244985
EMBL
NWSH01000578
PCG75534.1
PCG75533.1
KZ150249
PZC71833.1
BABH01025721
+ More
RSAL01000045 RVE50590.1 KQ458671 KPJ05556.1 AGBW02009205 OWR51385.1 ODYU01008354 SOQ51879.1 AK401676 BAM18298.1 JTDY01008796 KOB64391.1 JTDY01013133 KOB52184.1 GAMC01000249 JAC06307.1 CVRI01000057 CRL02306.1 CH933812 EDW05869.1 GAKP01022695 JAC36257.1 GBXI01007325 JAD06967.1 GDHF01016109 JAI36205.1 KRG07064.1 JRES01000520 KNC30450.1 CCAG010007469 CH902621 KPU81616.1 PYGN01000070 PSN55412.1 JXJN01015632 JXJN01015633 CH964291 KRG00376.1 AAAB01008811 EDO64480.2 ATLV01023319 KE525342 KFB48884.1 GFDF01009817 JAV04267.1 NEVH01000610 PNF43395.1 APCN01004772 AJWK01010859 CP012528 ALC48482.1 EDV44485.1 KK853497 KDR07203.1 EDW86374.2 AE014298 BT081991 ABW09343.1 ACO56230.1 KPU81617.1 CM000162 KRK05867.1 CH954180 KQS30460.1 GL453369 EFN76105.1 AXCN02001427 CH379064 KRT07001.1 OUUW01000011 SPP86330.1 KQ414864 KOC59992.1 CH940655 EDW66251.2 KQ981276 KYN43563.1 GL888067 EGI68280.1 KQ976444 KYM86205.1 ADTU01004475 EGK96283.1 QOIP01000003 RLU24515.1 KQ982720 KYQ51684.1 GEBQ01014299 JAT25678.1 KQ435782 KOX74757.1 GBYB01012420 JAG82187.1 AAF46134.2 KRT07003.1 SPP86331.1 SPP86328.1 KRT07002.1 EAL32150.3 GBYB01012421 JAG82188.1 GGMS01010485 MBY79688.1 GGMR01019665 MBY32284.1 KRK05866.1 KQS30457.1 KRT06999.1 SPP86329.1 GEDC01016780 JAS20518.1 JXUM01082443 JXUM01082444 JXUM01082445 GAPW01002689 KQ563313 JAC10909.1 KXJ74079.1 KZ288230 PBC31619.1 ABLF02029226 GL438128 EFN69543.1 CH480819 EDW52969.1 AHN59406.1
RSAL01000045 RVE50590.1 KQ458671 KPJ05556.1 AGBW02009205 OWR51385.1 ODYU01008354 SOQ51879.1 AK401676 BAM18298.1 JTDY01008796 KOB64391.1 JTDY01013133 KOB52184.1 GAMC01000249 JAC06307.1 CVRI01000057 CRL02306.1 CH933812 EDW05869.1 GAKP01022695 JAC36257.1 GBXI01007325 JAD06967.1 GDHF01016109 JAI36205.1 KRG07064.1 JRES01000520 KNC30450.1 CCAG010007469 CH902621 KPU81616.1 PYGN01000070 PSN55412.1 JXJN01015632 JXJN01015633 CH964291 KRG00376.1 AAAB01008811 EDO64480.2 ATLV01023319 KE525342 KFB48884.1 GFDF01009817 JAV04267.1 NEVH01000610 PNF43395.1 APCN01004772 AJWK01010859 CP012528 ALC48482.1 EDV44485.1 KK853497 KDR07203.1 EDW86374.2 AE014298 BT081991 ABW09343.1 ACO56230.1 KPU81617.1 CM000162 KRK05867.1 CH954180 KQS30460.1 GL453369 EFN76105.1 AXCN02001427 CH379064 KRT07001.1 OUUW01000011 SPP86330.1 KQ414864 KOC59992.1 CH940655 EDW66251.2 KQ981276 KYN43563.1 GL888067 EGI68280.1 KQ976444 KYM86205.1 ADTU01004475 EGK96283.1 QOIP01000003 RLU24515.1 KQ982720 KYQ51684.1 GEBQ01014299 JAT25678.1 KQ435782 KOX74757.1 GBYB01012420 JAG82187.1 AAF46134.2 KRT07003.1 SPP86331.1 SPP86328.1 KRT07002.1 EAL32150.3 GBYB01012421 JAG82188.1 GGMS01010485 MBY79688.1 GGMR01019665 MBY32284.1 KRK05866.1 KQS30457.1 KRT06999.1 SPP86329.1 GEDC01016780 JAS20518.1 JXUM01082443 JXUM01082444 JXUM01082445 GAPW01002689 KQ563313 JAC10909.1 KXJ74079.1 KZ288230 PBC31619.1 ABLF02029226 GL438128 EFN69543.1 CH480819 EDW52969.1 AHN59406.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000183832 UP000009192 UP000095301 UP000037069 UP000092444 UP000007801 UP000245037 UP000075880 UP000092460 UP000092443 UP000007798 UP000007062 UP000030765 UP000078200 UP000092445 UP000235965 UP000075903 UP000076407 UP000075840 UP000091820 UP000075902 UP000092461 UP000092553 UP000027135 UP000095300 UP000000803 UP000075885 UP000002282 UP000075881 UP000008711 UP000008237 UP000075886 UP000075920 UP000192221 UP000075884 UP000005203 UP000001819 UP000268350 UP000053825 UP000008792 UP000078541 UP000007755 UP000078540 UP000005205 UP000279307 UP000075809 UP000053105 UP000079169 UP000069272 UP000075900 UP000069940 UP000249989 UP000242457 UP000007819 UP000076408 UP000000311 UP000001292
UP000183832 UP000009192 UP000095301 UP000037069 UP000092444 UP000007801 UP000245037 UP000075880 UP000092460 UP000092443 UP000007798 UP000007062 UP000030765 UP000078200 UP000092445 UP000235965 UP000075903 UP000076407 UP000075840 UP000091820 UP000075902 UP000092461 UP000092553 UP000027135 UP000095300 UP000000803 UP000075885 UP000002282 UP000075881 UP000008711 UP000008237 UP000075886 UP000075920 UP000192221 UP000075884 UP000005203 UP000001819 UP000268350 UP000053825 UP000008792 UP000078541 UP000007755 UP000078540 UP000005205 UP000279307 UP000075809 UP000053105 UP000079169 UP000069272 UP000075900 UP000069940 UP000249989 UP000242457 UP000007819 UP000076408 UP000000311 UP000001292
Pfam
PF00587 tRNA-synt_2b
Interpro
SUPFAM
SSF54928
SSF54928
ProteinModelPortal
A0A2A4JV89
A0A2A4JVM7
A0A2W1BJJ6
H9J4X5
A0A3S2LCE5
A0A194QPX6
+ More
A0A212FCB2 A0A2H1WFL9 I4DK58 A0A0L7KMS4 A0A0L7K3E7 W8CBR0 A0A1J1IRE9 B4L673 A0A034V2A9 A0A0A1X766 A0A0K8VCF4 A0A1I8NF41 A0A0Q9XSK6 A0A0L0CDX7 A0A1B0FGI8 A0A0P9CFF5 A0A2P8ZFZ1 A0A182IR26 A0A1B0BJW1 A0A1A9YPL0 A0A0Q9X8G7 A7US28 A0A084WF90 A0A1A9VC71 A0A1B0A9W1 A0A1L8DD19 A0A2J7RRF9 A0A182USJ1 A0A182XNG1 A0A182IDQ9 A0A1A9WFW4 A0A182TGG9 A0A1B0CG90 A0A0M4F8T1 B3MQ59 A0A067QTL7 A0A1I8NZD9 B4NPD4 A8JUZ6 A0A0P9A5P1 A0A182PKY6 A0A0R1E9B2 A0A182K5G6 A0A0Q5T5B7 E2C7B8 A0A182QY71 A0A182VV53 A0A1W4VG14 A0A182NC44 A0A088AEH6 A0A0R3P1G5 A0A3B0KDJ3 A0A0L7QMW3 B4MAJ2 A0A195FSY9 F4WBZ1 A0A195BKY5 A0A158NZ15 F5HJ22 A0A3L8DWQ9 A0A1W4VSP0 A0A151WV06 A0A1B6LPR9 A0A0M9A0P0 A0A0C9RUC8 A0A3Q0JCK5 Q9W426 A0A182FMH7 A0A0R3P176 A0A3B0KMI6 A0A3B0KR09 A0A0R3P1A0 Q29GF9 A0A0C9RX14 A0A2S2QPM4 A0A2S2PSF8 A0A1W4VFQ8 A0A0R1EFH2 A0A0Q5T5B1 A0A0R3P173 A0A182RM50 A0A3B0KIR2 A0A1W4VTC4 A0A1B6D4C6 A0A023EQN0 A0A2A3EKB2 A0A1S4G7S3 J9KLL3 A0A182Y4T3 E2AAJ6 A0A1W4VU16 B4I0D8 X2JCR6
A0A212FCB2 A0A2H1WFL9 I4DK58 A0A0L7KMS4 A0A0L7K3E7 W8CBR0 A0A1J1IRE9 B4L673 A0A034V2A9 A0A0A1X766 A0A0K8VCF4 A0A1I8NF41 A0A0Q9XSK6 A0A0L0CDX7 A0A1B0FGI8 A0A0P9CFF5 A0A2P8ZFZ1 A0A182IR26 A0A1B0BJW1 A0A1A9YPL0 A0A0Q9X8G7 A7US28 A0A084WF90 A0A1A9VC71 A0A1B0A9W1 A0A1L8DD19 A0A2J7RRF9 A0A182USJ1 A0A182XNG1 A0A182IDQ9 A0A1A9WFW4 A0A182TGG9 A0A1B0CG90 A0A0M4F8T1 B3MQ59 A0A067QTL7 A0A1I8NZD9 B4NPD4 A8JUZ6 A0A0P9A5P1 A0A182PKY6 A0A0R1E9B2 A0A182K5G6 A0A0Q5T5B7 E2C7B8 A0A182QY71 A0A182VV53 A0A1W4VG14 A0A182NC44 A0A088AEH6 A0A0R3P1G5 A0A3B0KDJ3 A0A0L7QMW3 B4MAJ2 A0A195FSY9 F4WBZ1 A0A195BKY5 A0A158NZ15 F5HJ22 A0A3L8DWQ9 A0A1W4VSP0 A0A151WV06 A0A1B6LPR9 A0A0M9A0P0 A0A0C9RUC8 A0A3Q0JCK5 Q9W426 A0A182FMH7 A0A0R3P176 A0A3B0KMI6 A0A3B0KR09 A0A0R3P1A0 Q29GF9 A0A0C9RX14 A0A2S2QPM4 A0A2S2PSF8 A0A1W4VFQ8 A0A0R1EFH2 A0A0Q5T5B1 A0A0R3P173 A0A182RM50 A0A3B0KIR2 A0A1W4VTC4 A0A1B6D4C6 A0A023EQN0 A0A2A3EKB2 A0A1S4G7S3 J9KLL3 A0A182Y4T3 E2AAJ6 A0A1W4VU16 B4I0D8 X2JCR6
Ontologies
GO
PANTHER
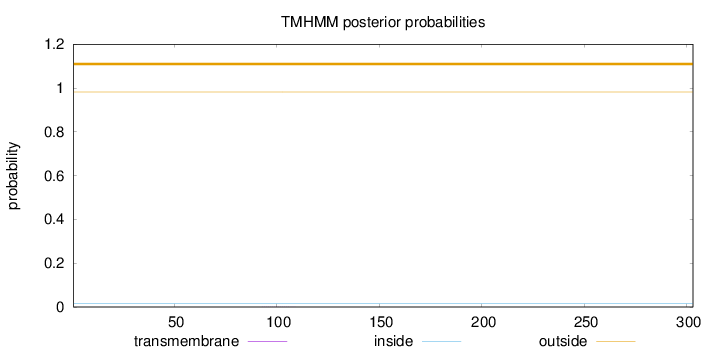
Topology
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00375
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01759
outside
1 - 303
Population Genetic Test Statistics
Pi
30.456435
Theta
32.314521
Tajima's D
-0.568624
CLR
0.005974
CSRT
0.228838558072096
Interpretation
Uncertain
