Gene
KWMTBOMO16494
Annotation
retroelement_polyprotein_[Lasius_niger]
Location in the cell
Nuclear Reliability : 3.92
Sequence
CDS
ATGGAGAAATTAAGATTTGATTTTGCAATGAAATCAATGGCGGATGGAAAAACAAACGTCATATGCATAACTTCCATTGCTACACCAAGTGGACAAGTTTTTGGAATTCCAGTCGAATATCAACCCGTAAATCTTCATAAGGTGATCACAACTACTCCTAACTACACTAGAGTAAAAAACTCTTTAGTAAAAACGCATCAAACGAGGAAAATATGGATAACTGTGACGGAGGAAATTTCAAAAACATATTTGGATGAAGAACAAAATTTGCAATTCAATGATTGTTATTTGGAAGAGATTTCAGAACAACCTGTTAGTTATGAATCGACATCTAGTGATTCTAATCACATGCTTGAAAAACTGTTAGAAAAACTACAAGAACAAAAACAGCAGAAACCAGAAAAACAAAATTTAGGGAAAATTGCAAAGGATTTTATTATAGAAAAATTCAACGGCCGTAATTCAAATGCATTACAGTGGATCAAAGATTTTAATAAAGAATGTGAACGTTTTCAAATAAACGAGGATAAACAAAAAATTGAAATTTTGAAGAACTTCTTAGAAAATGCTAGTGTAGATTGGTACAGTTGTATGCTGATGAAGCTTACAGTACAATCAGAATGGAGCAAATGGGAGAAAAAATTTTGTGAGACATTTGCGAACAAAGGATGGTCCTCCACTAGATATGCTCTTGCCTTTAAATATCAATCGGGATCCTTACTTGAGTATGCTTTAAAGAAGGAGAAATTATTGTTGGAAGTCAGGAAGACAGTGGATACCGGAACAATTATTGATCTCATAGCATTTGGTTTACCCAATTATGTGGCGGACAAGATTGACAAAGAAACATTACAAAGAACGGAAGACCTTTACAATGAGATAAGAAAATTGGAACATCTGGTGATGAAAAATAAATATGAAAAAAGGAACAGCTCATATAATTTAGATAATAAAGGGAAAAATATTGACGAAAAGAAACCATGCCCAATCTGTTTAAATGAGAAAAAAGGGAAACGGTTCCACCCAGAAGACAGCTGTTGGTTCAAAGAAAAAAATAAAAAAGCTGTGGTTTCTGGAATCTATGACTCTGGATCTAATGTTTCTTTAATTAATGCAAGATTATTGAAAATACAAGAGAAACAGAAGACAAATGATATGCAAGTTGTAAACTTAAAAACTATTAATGGTGAGAAAAAAACGAAAGGTATGGTAACGCTAAAAATAAAAATCTACAATATTGAAAGAAATATGGATATTTTTGTGGTGGATAATGAAAATTTTAACTATGACTTCTTAATTGGTTTGGATTGTATAAAAAAATTCAAATTAATACAAAATGAAGAACTTAACATCATACAATATCCCGAAAAGAAAAGCATAATCTCAAATGAACATTGTTTTAATGAGAATAATTTAACAAATGTCGGAATTCCTGATGTACTAGGTGACAAAAATAAAGAACAAACATTTAAAAAGAGAATAGAAGAGAAAGAGAAGATAGAGATGGAACTATACTCTGAGATAGTAAAAAAGGGTACTACTATAAATACCAACAAATTGTGTGAAATTAATTTCAATGAACATGTACACATTGATGAATTTCAAATGTCTGTAAATCATCTAGATTTATATCAGCAATCAAAAATTGAGAAGTTAGTAGATAAATACAAATCTGTTTTTGCTAAGGACAAATATGATGTTGGCTCTGTGCAGGATTATGAAGCCCATATAGACTTGATGGTAGATAAATACTGCTATAAGCGTCCGTACAGATGCACAATTGAAGACAGAACAGAAATTGAAAGTCAAATATCGAAGCTACTACAGAAAAATTTGATAGAGGAATCTTACAGCCCGTTTGCTGCTCCTGTAACTCTAGCATATAAGAAAGAAGATGATAAAAAAACAAGATTATGCATAGATTTCAGAGATTTAAACAAAATAGTAGTTCCACAATCACAACCTTTCCCACTTATAGAGGACTTAATGACCAAAACAGTAAACTGTAAGTATTATTCTACATTAGACATAAACTCAGCATTTTGGTCGATCCCATTGAAAATTCAAGATAGACATAAGACTGCTTTTGTGACACAAGAAGGACATTTTCAATGGACTTGTCTCCCATTTGGGTTAAAGACTGCACCAGCAATTTTCCAAAGAATATTGAGCAATATTATAAGAAAACACAGACTATCTAACTTTGCAGTAAACTTTATTGACGACATCTTAATTTTTTCTAAAACTTTTGAGGAACATATACAACACATATCGAGACTACTGGAAGCAATACAAAAGGAAGGGTTTAGACTGAAGTTTTCAAAATGTACATTTGCAAGTCACCATGCTAAATACTTGGGTCATATAATAGAAAACAATTCAGTTAGACCTCTGAAAGATTATTTAACTGCTGTCAAAAGCTTCCCAATTCCAGAAACAAGGAAAAATATACGGCAGTTTTTGGGAAAAGTAAATTTTCATCATAAGTTTGTACCACATAGTGCAATTATCCTCGATCCATTACATAACTTATTAAGAAAAGATGTGAAATTTGTATGGACGGCAGAATGCCAGGAAGCATTTGATAAAATTAGAGCATTGCTTTGCTCAGAACCAATCCTAAAAATATTTGACCCAGATTCACCGATAAGAATTTACACAGATGCTAGTATCAAAGGAGTTGGAGCAGTGTTGAAACAAGACGATGAAAACGGCGTGAGTAAACCGGTTGCTTATTTTTCGAAGAAATTAACAGAAGCACAGAAAAAGAAGAAGGCAATATACTTAGAATGTCTGGCGATAAAAGAAGCAGTCAAATACTGGCAACATTGGCTTATGGGAAAAGAATTTATGGTATACACAGATCATAAACCCCTGGAAAATATGAACATTAAAGCTCGAACTGATGAGGAACTAGCCAAAAATATTACAGCAAACATTAATAAAATGTGTGATGCGTGTGAAACCTGCATTAAAAACAAGTCCAGGAGGAAGTCGAAATATGGTTTAATGTCACATTTGGGACCAGCCACACACCCGTTTGAAATTGTATCCATCGATACTATTGGGGGCTTTGGAGGATCCCGATCAACCAAAAGCGAAGGTGTCCCGTCTGTCGCGCTCACGTTCAGCACGCGGGAGTCGGCTCGGATGTCGTTCAGATCGCAGGCGAGACTCACCACGCCCGTGCTCGAATCCAACGAGAAGCAATTATCATCATTGCCGCCGACGATTCGATAA
Protein
MEKLRFDFAMKSMADGKTNVICITSIATPSGQVFGIPVEYQPVNLHKVITTTPNYTRVKNSLVKTHQTRKIWITVTEEISKTYLDEEQNLQFNDCYLEEISEQPVSYESTSSDSNHMLEKLLEKLQEQKQQKPEKQNLGKIAKDFIIEKFNGRNSNALQWIKDFNKECERFQINEDKQKIEILKNFLENASVDWYSCMLMKLTVQSEWSKWEKKFCETFANKGWSSTRYALAFKYQSGSLLEYALKKEKLLLEVRKTVDTGTIIDLIAFGLPNYVADKIDKETLQRTEDLYNEIRKLEHLVMKNKYEKRNSSYNLDNKGKNIDEKKPCPICLNEKKGKRFHPEDSCWFKEKNKKAVVSGIYDSGSNVSLINARLLKIQEKQKTNDMQVVNLKTINGEKKTKGMVTLKIKIYNIERNMDIFVVDNENFNYDFLIGLDCIKKFKLIQNEELNIIQYPEKKSIISNEHCFNENNLTNVGIPDVLGDKNKEQTFKKRIEEKEKIEMELYSEIVKKGTTINTNKLCEINFNEHVHIDEFQMSVNHLDLYQQSKIEKLVDKYKSVFAKDKYDVGSVQDYEAHIDLMVDKYCYKRPYRCTIEDRTEIESQISKLLQKNLIEESYSPFAAPVTLAYKKEDDKKTRLCIDFRDLNKIVVPQSQPFPLIEDLMTKTVNCKYYSTLDINSAFWSIPLKIQDRHKTAFVTQEGHFQWTCLPFGLKTAPAIFQRILSNIIRKHRLSNFAVNFIDDILIFSKTFEEHIQHISRLLEAIQKEGFRLKFSKCTFASHHAKYLGHIIENNSVRPLKDYLTAVKSFPIPETRKNIRQFLGKVNFHHKFVPHSAIILDPLHNLLRKDVKFVWTAECQEAFDKIRALLCSEPILKIFDPDSPIRIYTDASIKGVGAVLKQDDENGVSKPVAYFSKKLTEAQKKKKAIYLECLAIKEAVKYWQHWLMGKEFMVYTDHKPLENMNIKARTDEELAKNITANINKMCDACETCIKNKSRRKSKYGLMSHLGPATHPFEIVSIDTIGGFGGSRSTKSEGVPSVALTFSTRESARMSFRSQARLTTPVLESNEKQLSSLPPTIR
Summary
Uniprot
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
PDB
4OL8
E-value=2.18258e-67,
Score=653
Ontologies
GO
Topology
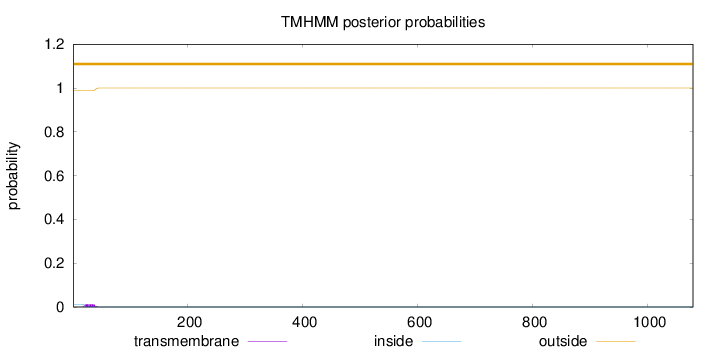
Length:
1079
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25361
Exp number, first 60 AAs:
0.25168
Total prob of N-in:
0.01254
outside
1 - 1079
Population Genetic Test Statistics
Pi
10.999681
Theta
10.461729
Tajima's D
-1.384835
CLR
0.938055
CSRT
0.072346382680866
Interpretation
Uncertain
