Pre Gene Modal
BGIBMGA005634
Annotation
PREDICTED:_protein_shuttle_craft_[Bombyx_mori]
Full name
Protein shuttle craft
Transcription factor
Location in the cell
Extracellular Reliability : 3.251
Sequence
CDS
ATGTGCAGCAGCAAGCTACCGCAAGTGTGCGGGCGTCCTTGCCAACGCATGCTACAGTGCGGCGTACACAAATGCATGAAGGAATGCCACGAGGGTCCCTGTGAAGACTGCACCGAGTCTGTTACACAAGTATGCCATTGCCCGCTGGCCAAGACTCGTGAGGTCGCGTGCCGCGTGGAGTCGTCGTGGTCGTGCGGCGGCGCGTGCGGGCGCGTGCTATCGTGCGGCGCGCACGTGTGCCGGAGCACGTGCCACGAGCCCCCCTGCCCCTCCTGCGACCTGCTGCCTGCCGCCCTCATCACATGTCCGTGCGGGAAGACTAGAATACCAAAAGACAAACGCAAGTCGTGCACGGATCCGATCCCGTTGTGCGGAAACATCTGCGCCAAGCCGCTGCCCTGCGGCCCGGCCGGTGACAAGCATTTCTGCAAACTAGACTGTCATGAAGGTCCTTGTCCCGTTTGTCCTGACAAGACCTTATTGCCGTGCCGCTGTGGTCATTCGAACCGCGAGGTTCCGTGCGCTGAGCTACCGGACATGTATAACAATGTGTTCTGTCAGAAGAAATGTAATAAGAAACTGTCGTGCGGTCGACACCGTTGCCGCGCGTGCTGCTGCGCGGACCCGCAGCACCGGTGCGGGCTCGTGTGCGGGCGCTCGCTCGCCTGCCAGACGCACCGCTGTCAGGACTTCTGCCACACCGGACACTGCGCGCCGTGCCCCACGCTCAGCTTCGAGGAGCTGAGCTGTACGTGCGGGCGCGAGGTGCTGCTGCCGCCCGTGCGCTGCGGCACGCGGCCCCCGCCCTGCAGCGCGCCGTGCCGCCGCGCGCGCCCCTGCGGGCACGAGCCGCTGCACTCGTGCCACACCGGGGACTGTCCGCCCTGCGTCGTGCTCACCACCAGGACCTGCCACGGCGGACACGAGGAACGAAAAACCATCCCGTGTTCGCAAGAAGAGTTCTCGTGCGGCCTCCCGTGCGGCAAACCTCTCCCTTGCGGTAAACATACCTGCATCAAGGTCTGTCACAAGGGACCTTGCGATGCCGGCAAGTGCACCCAGCCGTGCAACGAGAAGCGTGCGAGCTGCGGGCACCCGTGCGGCGCCGCGTGCCACTCCGGCGCGGCGGGCGGCGCGGGCGCGCTGTGCCCGAGCGCGGCGCCGTGCCGCCGCACCGTGCGCGCCACGTGCCCCTGCAAACGGAAATCAGCCGAGAGACCTTGCTTCGAAAACGCACGGGATTACGATAAAATGATGAGTGCTCTTGCCGCCACCAAGATTCAGGAGGGAGGCTCTGTTGACTTGTCCGACGCACAGCGCGCCGTCAATATGCTCAAAACGCTGGAGTGCGACGACGAGTGCCGCGTGGAGGCGCGCACGCGGCAGATGGCGCTGGCGCTGCAGATCCGCAACCCGGACGTGTCCGCCAAGCTGGCGCCGCGCTACAGCGAGCACGTGCGCGCCACCGCCCAGCGCGAGCCCGCCTTCGCCTACCACGTGCACAACCAGCTCACCGAGCTCGTGCACCTCGCCAAGAAGTCGAAACAGAAGACGCGCTCGCACTCTTTCCCGTCGATGAACAGACAGAAGCGGCAGTTCATCCACGAGCTGTGCGAGCACTTCGGCTGCGAGAGCGTGGCCTACGACGCGGAGCCCAACAGGAACGTGGTGGCCACTGCCGACAAGGAGAAGTCGTGGTTGCCGGCGATGAGCATAATGGAAGTGTTGTCGCGTGAGGCGGGCCGGCGGCGCGTGCCCGGCCCCGTGCTGCGGACCGCGCACTCCTCCGCCGCCTCTTCAGCCTCGACGACCTCCTCCTCCGCCGCTAACAAGTAA
Protein
MCSSKLPQVCGRPCQRMLQCGVHKCMKECHEGPCEDCTESVTQVCHCPLAKTREVACRVESSWSCGGACGRVLSCGAHVCRSTCHEPPCPSCDLLPAALITCPCGKTRIPKDKRKSCTDPIPLCGNICAKPLPCGPAGDKHFCKLDCHEGPCPVCPDKTLLPCRCGHSNREVPCAELPDMYNNVFCQKKCNKKLSCGRHRCRACCCADPQHRCGLVCGRSLACQTHRCQDFCHTGHCAPCPTLSFEELSCTCGREVLLPPVRCGTRPPPCSAPCRRARPCGHEPLHSCHTGDCPPCVVLTTRTCHGGHEERKTIPCSQEEFSCGLPCGKPLPCGKHTCIKVCHKGPCDAGKCTQPCNEKRASCGHPCGAACHSGAAGGAGALCPSAAPCRRTVRATCPCKRKSAERPCFENARDYDKMMSALAATKIQEGGSVDLSDAQRAVNMLKTLECDDECRVEARTRQMALALQIRNPDVSAKLAPRYSEHVRATAQREPAFAYHVHNQLTELVHLAKKSKQKTRSHSFPSMNRQKRQFIHELCEHFGCESVAYDAEPNRNVVATADKEKSWLPAMSIMEVLSREAGRRRVPGPVLRTAHSSAASSASTTSSSAANK
Summary
Similarity
Belongs to the NFX1 family.
Keywords
Alternative splicing
Complete proteome
DNA-binding
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
RNA-binding
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Protein shuttle craft
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J7Z2
A0A2A4K355
A0A2A4K2T6
A0A3S2LJM5
A0A2H1V9F9
A0A194QGR9
+ More
A0A194QXJ0 A0A212EP74 L7X1P9 D0AB98 A0A2P8Z3T6 D6WMN1 A0A139WHD1 A0A2J7QGB1 A0A2J7QGA5 A0A1B6EVN5 A0A1B6JPB1 E0VEM9 A0A1B6KND9 A0A2A3E3N7 A0A088ART8 A0A067R6D8 W8APS1 A0A026WYJ9 A0A195C826 A0A1Y1JZ76 E2C3V1 F4WM20 V5GY42 A0A0L7RDI2 A0A0L0CJR7 A0A1B0GP93 A0A195FUE9 A0A154PBD9 B4MZE5 A0A0M4E2B7 A0A1I8Q8G4 E8NH79 A0A034VNI7 A0A0P4VKR5 A0A0A1X737 A0A0V0G909 A0A224XHM8 A0A069DY12 T1P9M2 A0A1Q3FEQ8 B3MM96 A0A1Q3FEQ0 A0A1Q3FCB1 A0A1I8N6H3 A0A1I8N6G0 B4NYD7 A0A1W4WC55 A0A0A9ZGP6 A0A1B0A839 A0A1B0FP18 A0A146LIZ1 A0A3B0KT64 A0A1A9V167 Q29P77 B3N5P4 B4KLB0 B4GK89 A0A1A9X8Y5 B4LRX1 A0A0M3QTY5 A0A1B0BFJ2 A0A1B0CN35 A0A3B0L0K6 A0A1A9X299 M9PDG3 P40798-2 A0A310SMP3 M9PD57 B0W1A0 P40798 A0A182KWI0 A0A182MHE8 A0A0K8V596 A0A0J9R264 A0A0J9R2N5 Q17KM1 B4HXM6 T1IAW5 E9H0P6 W5JVC9 A0A2M4AEV2 A0A182F3S3 B4JAX9 A0A023EY35 A0A0P5VH86 A0A182PVX0 A0A0P5YM02 B4Q602 A0A0P5XCR0 A0A182H838 A0A0P5A3Q8 B4LWS2 A0A0P5AUP1 A0A084VMJ6
A0A194QXJ0 A0A212EP74 L7X1P9 D0AB98 A0A2P8Z3T6 D6WMN1 A0A139WHD1 A0A2J7QGB1 A0A2J7QGA5 A0A1B6EVN5 A0A1B6JPB1 E0VEM9 A0A1B6KND9 A0A2A3E3N7 A0A088ART8 A0A067R6D8 W8APS1 A0A026WYJ9 A0A195C826 A0A1Y1JZ76 E2C3V1 F4WM20 V5GY42 A0A0L7RDI2 A0A0L0CJR7 A0A1B0GP93 A0A195FUE9 A0A154PBD9 B4MZE5 A0A0M4E2B7 A0A1I8Q8G4 E8NH79 A0A034VNI7 A0A0P4VKR5 A0A0A1X737 A0A0V0G909 A0A224XHM8 A0A069DY12 T1P9M2 A0A1Q3FEQ8 B3MM96 A0A1Q3FEQ0 A0A1Q3FCB1 A0A1I8N6H3 A0A1I8N6G0 B4NYD7 A0A1W4WC55 A0A0A9ZGP6 A0A1B0A839 A0A1B0FP18 A0A146LIZ1 A0A3B0KT64 A0A1A9V167 Q29P77 B3N5P4 B4KLB0 B4GK89 A0A1A9X8Y5 B4LRX1 A0A0M3QTY5 A0A1B0BFJ2 A0A1B0CN35 A0A3B0L0K6 A0A1A9X299 M9PDG3 P40798-2 A0A310SMP3 M9PD57 B0W1A0 P40798 A0A182KWI0 A0A182MHE8 A0A0K8V596 A0A0J9R264 A0A0J9R2N5 Q17KM1 B4HXM6 T1IAW5 E9H0P6 W5JVC9 A0A2M4AEV2 A0A182F3S3 B4JAX9 A0A023EY35 A0A0P5VH86 A0A182PVX0 A0A0P5YM02 B4Q602 A0A0P5XCR0 A0A182H838 A0A0P5A3Q8 B4LWS2 A0A0P5AUP1 A0A084VMJ6
Pubmed
19121390
26354079
22118469
23674305
29403074
18362917
+ More
19820115 20566863 24845553 24495485 24508170 28004739 20798317 21719571 26108605 17994087 25348373 27129103 25830018 26334808 25315136 17550304 25401762 26823975 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8524296 10471707 10731138 18327897 20966253 22936249 17510324 21292972 20920257 23761445 24945155 26483478 24438588
19820115 20566863 24845553 24495485 24508170 28004739 20798317 21719571 26108605 17994087 25348373 27129103 25830018 26334808 25315136 17550304 25401762 26823975 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8524296 10471707 10731138 18327897 20966253 22936249 17510324 21292972 20920257 23761445 24945155 26483478 24438588
EMBL
BABH01020191
BABH01020192
NWSH01000227
PCG78213.1
PCG78214.1
RSAL01000090
+ More
RVE48091.1 ODYU01001380 SOQ37468.1 KQ459232 KPJ02641.1 KQ461155 KPJ08291.1 AGBW02013517 OWR43279.1 KC469893 AGC92703.2 FP102340 CBH09293.1 PYGN01000208 PSN51156.1 KQ971343 EFA04758.2 KYB27326.1 NEVH01014379 PNF27624.1 PNF27625.1 GECZ01027838 JAS41931.1 GECU01009311 GECU01006610 JAS98395.1 JAT01097.1 DS235093 EEB11835.1 GEBQ01027027 JAT12950.1 KZ288427 PBC25866.1 KK852714 KDR17911.1 GAMC01020057 GAMC01020055 JAB86498.1 KK107064 EZA60846.1 KQ978220 KYM96321.1 GEZM01096724 JAV54622.1 GL452364 EFN77337.1 GL888217 EGI64664.1 GALX01003148 JAB65318.1 KQ414614 KOC68840.1 JRES01000305 KNC32491.1 AJVK01033236 AJVK01033237 KQ981264 KYN44046.1 KQ434856 KZC08714.1 CH963913 EDW77484.2 CP012523 ALC39716.1 BT125955 ADX35934.1 GAKP01015839 JAC43113.1 GDKW01003695 JAI52900.1 GBXI01007360 JAD06932.1 GECL01001450 JAP04674.1 GFTR01008466 JAW07960.1 GBGD01000277 JAC88612.1 KA645427 AFP60056.1 GFDL01009011 JAV26034.1 CH902620 EDV31856.1 GFDL01009025 JAV26020.1 GFDL01009855 JAV25190.1 CM000157 EDW89773.1 GBHO01007718 GBHO01000278 JAG35886.1 JAG43326.1 CCAG010018380 GDHC01011747 JAQ06882.1 OUUW01000040 SPP89929.1 CH379058 EAL34416.2 CH954177 EDV58003.1 CH933807 EDW11771.1 CH479184 EDW37055.1 CH940649 EDW63647.1 ALC39723.1 JXJN01013499 AJWK01019755 SPP89928.1 AE014134 AGB92995.1 U09306 AF145679 KQ760869 OAD58748.1 AGB92996.1 DS231821 EDS44970.1 AXCM01000660 GDHF01018212 GDHF01005212 JAI34102.1 JAI47102.1 CM002910 KMY90342.1 KMY90343.1 CH477223 EAT47205.1 CH480818 EDW51806.1 ACPB03000855 GL732581 EFX74683.1 ADMH02000386 ETN66754.1 GGFK01006000 MBW39321.1 CH916368 EDW02849.1 GAPW01000154 JAC13444.1 GDIP01115026 JAL88688.1 GDIP01055985 JAM47730.1 CM000361 EDX05105.1 GDIP01074037 JAM29678.1 JXUM01028608 JXUM01028609 JXUM01028610 KQ560826 KXJ80724.1 GDIP01207964 JAJ15438.1 CH940650 EDW66643.2 GDIP01208014 JAJ15388.1 ATLV01014599 KE524975 KFB39190.1
RVE48091.1 ODYU01001380 SOQ37468.1 KQ459232 KPJ02641.1 KQ461155 KPJ08291.1 AGBW02013517 OWR43279.1 KC469893 AGC92703.2 FP102340 CBH09293.1 PYGN01000208 PSN51156.1 KQ971343 EFA04758.2 KYB27326.1 NEVH01014379 PNF27624.1 PNF27625.1 GECZ01027838 JAS41931.1 GECU01009311 GECU01006610 JAS98395.1 JAT01097.1 DS235093 EEB11835.1 GEBQ01027027 JAT12950.1 KZ288427 PBC25866.1 KK852714 KDR17911.1 GAMC01020057 GAMC01020055 JAB86498.1 KK107064 EZA60846.1 KQ978220 KYM96321.1 GEZM01096724 JAV54622.1 GL452364 EFN77337.1 GL888217 EGI64664.1 GALX01003148 JAB65318.1 KQ414614 KOC68840.1 JRES01000305 KNC32491.1 AJVK01033236 AJVK01033237 KQ981264 KYN44046.1 KQ434856 KZC08714.1 CH963913 EDW77484.2 CP012523 ALC39716.1 BT125955 ADX35934.1 GAKP01015839 JAC43113.1 GDKW01003695 JAI52900.1 GBXI01007360 JAD06932.1 GECL01001450 JAP04674.1 GFTR01008466 JAW07960.1 GBGD01000277 JAC88612.1 KA645427 AFP60056.1 GFDL01009011 JAV26034.1 CH902620 EDV31856.1 GFDL01009025 JAV26020.1 GFDL01009855 JAV25190.1 CM000157 EDW89773.1 GBHO01007718 GBHO01000278 JAG35886.1 JAG43326.1 CCAG010018380 GDHC01011747 JAQ06882.1 OUUW01000040 SPP89929.1 CH379058 EAL34416.2 CH954177 EDV58003.1 CH933807 EDW11771.1 CH479184 EDW37055.1 CH940649 EDW63647.1 ALC39723.1 JXJN01013499 AJWK01019755 SPP89928.1 AE014134 AGB92995.1 U09306 AF145679 KQ760869 OAD58748.1 AGB92996.1 DS231821 EDS44970.1 AXCM01000660 GDHF01018212 GDHF01005212 JAI34102.1 JAI47102.1 CM002910 KMY90342.1 KMY90343.1 CH477223 EAT47205.1 CH480818 EDW51806.1 ACPB03000855 GL732581 EFX74683.1 ADMH02000386 ETN66754.1 GGFK01006000 MBW39321.1 CH916368 EDW02849.1 GAPW01000154 JAC13444.1 GDIP01115026 JAL88688.1 GDIP01055985 JAM47730.1 CM000361 EDX05105.1 GDIP01074037 JAM29678.1 JXUM01028608 JXUM01028609 JXUM01028610 KQ560826 KXJ80724.1 GDIP01207964 JAJ15438.1 CH940650 EDW66643.2 GDIP01208014 JAJ15388.1 ATLV01014599 KE524975 KFB39190.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000245037 UP000007266 UP000235965 UP000009046 UP000242457 UP000005203 UP000027135 UP000053097 UP000078542 UP000008237 UP000007755 UP000053825 UP000037069 UP000092462 UP000078541 UP000076502 UP000007798 UP000092553 UP000095300 UP000007801 UP000095301 UP000002282 UP000192221 UP000092445 UP000092444 UP000268350 UP000078200 UP000001819 UP000008711 UP000009192 UP000008744 UP000092443 UP000008792 UP000092460 UP000092461 UP000091820 UP000000803 UP000002320 UP000075882 UP000075883 UP000008820 UP000001292 UP000015103 UP000000305 UP000000673 UP000069272 UP000001070 UP000075885 UP000000304 UP000069940 UP000249989 UP000030765
UP000245037 UP000007266 UP000235965 UP000009046 UP000242457 UP000005203 UP000027135 UP000053097 UP000078542 UP000008237 UP000007755 UP000053825 UP000037069 UP000092462 UP000078541 UP000076502 UP000007798 UP000092553 UP000095300 UP000007801 UP000095301 UP000002282 UP000192221 UP000092445 UP000092444 UP000268350 UP000078200 UP000001819 UP000008711 UP000009192 UP000008744 UP000092443 UP000008792 UP000092460 UP000092461 UP000091820 UP000000803 UP000002320 UP000075882 UP000075883 UP000008820 UP000001292 UP000015103 UP000000305 UP000000673 UP000069272 UP000001070 UP000075885 UP000000304 UP000069940 UP000249989 UP000030765
Interpro
SUPFAM
SSF82708
SSF82708
Gene 3D
ProteinModelPortal
H9J7Z2
A0A2A4K355
A0A2A4K2T6
A0A3S2LJM5
A0A2H1V9F9
A0A194QGR9
+ More
A0A194QXJ0 A0A212EP74 L7X1P9 D0AB98 A0A2P8Z3T6 D6WMN1 A0A139WHD1 A0A2J7QGB1 A0A2J7QGA5 A0A1B6EVN5 A0A1B6JPB1 E0VEM9 A0A1B6KND9 A0A2A3E3N7 A0A088ART8 A0A067R6D8 W8APS1 A0A026WYJ9 A0A195C826 A0A1Y1JZ76 E2C3V1 F4WM20 V5GY42 A0A0L7RDI2 A0A0L0CJR7 A0A1B0GP93 A0A195FUE9 A0A154PBD9 B4MZE5 A0A0M4E2B7 A0A1I8Q8G4 E8NH79 A0A034VNI7 A0A0P4VKR5 A0A0A1X737 A0A0V0G909 A0A224XHM8 A0A069DY12 T1P9M2 A0A1Q3FEQ8 B3MM96 A0A1Q3FEQ0 A0A1Q3FCB1 A0A1I8N6H3 A0A1I8N6G0 B4NYD7 A0A1W4WC55 A0A0A9ZGP6 A0A1B0A839 A0A1B0FP18 A0A146LIZ1 A0A3B0KT64 A0A1A9V167 Q29P77 B3N5P4 B4KLB0 B4GK89 A0A1A9X8Y5 B4LRX1 A0A0M3QTY5 A0A1B0BFJ2 A0A1B0CN35 A0A3B0L0K6 A0A1A9X299 M9PDG3 P40798-2 A0A310SMP3 M9PD57 B0W1A0 P40798 A0A182KWI0 A0A182MHE8 A0A0K8V596 A0A0J9R264 A0A0J9R2N5 Q17KM1 B4HXM6 T1IAW5 E9H0P6 W5JVC9 A0A2M4AEV2 A0A182F3S3 B4JAX9 A0A023EY35 A0A0P5VH86 A0A182PVX0 A0A0P5YM02 B4Q602 A0A0P5XCR0 A0A182H838 A0A0P5A3Q8 B4LWS2 A0A0P5AUP1 A0A084VMJ6
A0A194QXJ0 A0A212EP74 L7X1P9 D0AB98 A0A2P8Z3T6 D6WMN1 A0A139WHD1 A0A2J7QGB1 A0A2J7QGA5 A0A1B6EVN5 A0A1B6JPB1 E0VEM9 A0A1B6KND9 A0A2A3E3N7 A0A088ART8 A0A067R6D8 W8APS1 A0A026WYJ9 A0A195C826 A0A1Y1JZ76 E2C3V1 F4WM20 V5GY42 A0A0L7RDI2 A0A0L0CJR7 A0A1B0GP93 A0A195FUE9 A0A154PBD9 B4MZE5 A0A0M4E2B7 A0A1I8Q8G4 E8NH79 A0A034VNI7 A0A0P4VKR5 A0A0A1X737 A0A0V0G909 A0A224XHM8 A0A069DY12 T1P9M2 A0A1Q3FEQ8 B3MM96 A0A1Q3FEQ0 A0A1Q3FCB1 A0A1I8N6H3 A0A1I8N6G0 B4NYD7 A0A1W4WC55 A0A0A9ZGP6 A0A1B0A839 A0A1B0FP18 A0A146LIZ1 A0A3B0KT64 A0A1A9V167 Q29P77 B3N5P4 B4KLB0 B4GK89 A0A1A9X8Y5 B4LRX1 A0A0M3QTY5 A0A1B0BFJ2 A0A1B0CN35 A0A3B0L0K6 A0A1A9X299 M9PDG3 P40798-2 A0A310SMP3 M9PD57 B0W1A0 P40798 A0A182KWI0 A0A182MHE8 A0A0K8V596 A0A0J9R264 A0A0J9R2N5 Q17KM1 B4HXM6 T1IAW5 E9H0P6 W5JVC9 A0A2M4AEV2 A0A182F3S3 B4JAX9 A0A023EY35 A0A0P5VH86 A0A182PVX0 A0A0P5YM02 B4Q602 A0A0P5XCR0 A0A182H838 A0A0P5A3Q8 B4LWS2 A0A0P5AUP1 A0A084VMJ6
Ontologies
GO
PANTHER
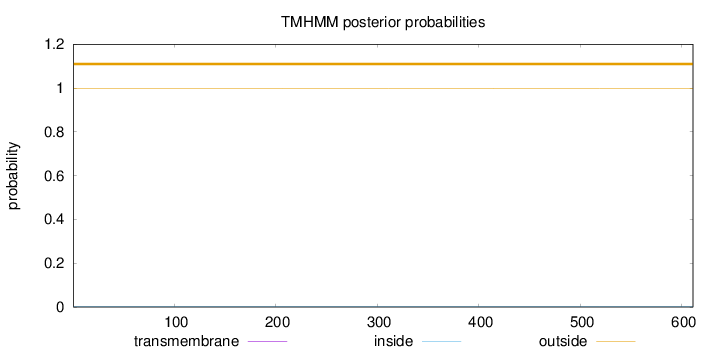
Topology
Subcellular location
Nucleus
Length:
611
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00063
outside
1 - 611
Population Genetic Test Statistics
Pi
24.776219
Theta
36.513728
Tajima's D
-0.979608
CLR
0.012208
CSRT
0.140742962851857
Interpretation
Uncertain
