Pre Gene Modal
BGIBMGA005634
Annotation
PREDICTED:_protein_shuttle_craft_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.358
Sequence
CDS
ATGTCGCAGTGGAACAACTCTTACTCCTACAATGGTCAATACCAAGCCAACGGCTGGAACCGAGATGTTAATGGCCAATATTCAAATTCACAGTATTATAATAACAGGCAAGATTCAAATAATCAATATGTGAGCTTCAATGAGTTCTTAACTCAAATACAAGGCGACAGTGCATCAGTCCCGGGCAATTCGGTTCATTATAATAATGTAAGATATGAAAACTATCCCACCACTCAATACAATTATCAAAACATGCCGTCTACTTCACAGAATAGATCACTAGATAGTTACAATTATCCAACAAATGTGAACCCAAGGAATCCCCAACAGTACCAAATGAATATGCAGAATCAATATGTGCCATCTGAAAGCTATAATAATGAAATTATACTAAAATCTAACTTAACACCTACTGCAACAGAATTTGTACCAAAGAGTATGGCTAAACAGATCACTGGTCACCAAAGTTTAGCAGAACCTGCAACTAATAACGTTAAGCATAATGATGCAAACTGGAGAGAAAGAGAGAGTTTACAACAAAATGGTGAAATTGTAACTAATCAGGTTTCTGAAAATAACAAAAGCCAATACTCCAAAACTCAAAACAGGGTACCTAATAGCCAAGATAATGCGCATTCAAGTGAATCCAATGTTAAAGTTAACGAACCAAGGCACAGAAATGGACATCGTAACAATGAGGCTAATGGTAGTTACTACAATCAAAATAATCAAAGTGAAAGTAAATCTCACAGTAATGAAATAAATAACCATAGTGATTCAAGTAGTAGATACTATGAAAATCAAAGAAATTATGATAGGAATGAGAAGAAAATCATATCAAAGACGAATCCCAAATCAAAACCAAGAGATGCAGATGCTCGTACATTCTACAACAGTTCCATTACTAAGGACAGTCAAGATGTTCGGAATGGAAGAGGTGAGAGTTCTGGCAGGCAAAGGAATTGGGCTGGAAGTCAAAGAATCAAATCTTCAGAGAAGTCAACTCCTGAAGATGAGCAATATGCTACCTCATACACTCAACAGAAACAGGAGAAAGTAGAGAGAAATAAAACAGAGAACATTCCAAGCCCAAACAGGTTTAGAAGCAAAAATATTGAACAAGCAATAAACAAAGAAATGACACAAAGGGAACGCCTGACTGATCAATTAGAAAAAGGAACATTAGAGTGTCTAGTATGTTGCGAGAGAGTTAAACAAACAGAATCCGTGTGGTCGTGTGGCAACTGCTACCATGTACTCCATCTTAGGTGCATAAGAAAATGGGCAATGAGTAGTTTGGTCGGTGAGTTCAATTAA
Protein
MSQWNNSYSYNGQYQANGWNRDVNGQYSNSQYYNNRQDSNNQYVSFNEFLTQIQGDSASVPGNSVHYNNVRYENYPTTQYNYQNMPSTSQNRSLDSYNYPTNVNPRNPQQYQMNMQNQYVPSESYNNEIILKSNLTPTATEFVPKSMAKQITGHQSLAEPATNNVKHNDANWRERESLQQNGEIVTNQVSENNKSQYSKTQNRVPNSQDNAHSSESNVKVNEPRHRNGHRNNEANGSYYNQNNQSESKSHSNEINNHSDSSSRYYENQRNYDRNEKKIISKTNPKSKPRDADARTFYNSSITKDSQDVRNGRGESSGRQRNWAGSQRIKSSEKSTPEDEQYATSYTQQKQEKVERNKTENIPSPNRFRSKNIEQAINKEMTQRERLTDQLEKGTLECLVCCERVKQTESVWSCGNCYHVLHLRCIRKWAMSSLVGEFN
Summary
Uniprot
H9J7Z2
A0A194QXJ0
A0A194QGR9
A0A3S2LJM5
A0A1E1WDV1
A0A212EP74
+ More
D0AB98 A0A2A4K2T6 A0A2A4K355 L7X1P9 A0A2H1V9F9 G3KAV1 G3KAS7 G3KAR4 G3KAR1 G3KAS0 G3KAR2 G3KAS9 G3KAT2 G3KAU5 G3KAS1 G3KAU1 G3KAS8 G3KAR0 G3KAV0 G3KAU0 G3KAQ7 G3KAS2 G3KAU7 G3KAT7 G3KAR7 G3KAU4 G3KAT1 G3KAU3 G3KAU8 G3KAT9 G3KAS5 G3KAS4 G3KAT0 G3KAQ9 G3KAT8 G3KAQ6 G3KAT5 G3KAQ8 G3KAR6 G3KAU2 G3KAR9 G3KAS6 G3KAR8 G3KAR5 G3KAS3 A0A1B0GP93 E0VEM9
D0AB98 A0A2A4K2T6 A0A2A4K355 L7X1P9 A0A2H1V9F9 G3KAV1 G3KAS7 G3KAR4 G3KAR1 G3KAS0 G3KAR2 G3KAS9 G3KAT2 G3KAU5 G3KAS1 G3KAU1 G3KAS8 G3KAR0 G3KAV0 G3KAU0 G3KAQ7 G3KAS2 G3KAU7 G3KAT7 G3KAR7 G3KAU4 G3KAT1 G3KAU3 G3KAU8 G3KAT9 G3KAS5 G3KAS4 G3KAT0 G3KAQ9 G3KAT8 G3KAQ6 G3KAT5 G3KAQ8 G3KAR6 G3KAU2 G3KAR9 G3KAS6 G3KAR8 G3KAR5 G3KAS3 A0A1B0GP93 E0VEM9
EMBL
BABH01020191
BABH01020192
KQ461155
KPJ08291.1
KQ459232
KPJ02641.1
+ More
RSAL01000090 RVE48091.1 GDQN01005891 JAT85163.1 AGBW02013517 OWR43279.1 FP102340 CBH09293.1 NWSH01000227 PCG78214.1 PCG78213.1 KC469893 AGC92703.2 ODYU01001380 SOQ37468.1 JN175123 AEN68466.1 JN175099 AEN68442.1 JN175086 AEN68429.1 JN175083 AEN68426.1 JN175092 AEN68435.1 JN175084 AEN68427.1 JN175101 AEN68444.1 JN175104 AEN68447.1 JN175117 AEN68460.1 JN175085 JN175093 AEN68428.1 AEN68436.1 JN175113 AEN68456.1 JN175100 AEN68443.1 JN175082 AEN68425.1 JN175122 AEN68465.1 JN175112 AEN68455.1 JN175079 AEN68422.1 JN175094 AEN68437.1 JN175106 JN175118 JN175119 AEN68449.1 AEN68461.1 AEN68462.1 JN175109 AEN68452.1 JN175089 AEN68432.1 JN175116 AEN68459.1 JN175103 AEN68446.1 JN175115 AEN68458.1 JN175120 AEN68463.1 JN175111 AEN68454.1 JN175097 AEN68440.1 JN175096 AEN68439.1 JN175102 AEN68445.1 JN175081 AEN68424.1 JN175110 JN175121 AEN68453.1 AEN68464.1 JN175078 AEN68421.1 JN175105 JN175107 JN175108 AEN68448.1 AEN68450.1 AEN68451.1 JN175080 AEN68423.1 JN175088 AEN68431.1 JN175114 AEN68457.1 JN175091 AEN68434.1 JN175098 AEN68441.1 JN175090 AEN68433.1 JN175087 AEN68430.1 JN175095 AEN68438.1 AJVK01033236 AJVK01033237 DS235093 EEB11835.1
RSAL01000090 RVE48091.1 GDQN01005891 JAT85163.1 AGBW02013517 OWR43279.1 FP102340 CBH09293.1 NWSH01000227 PCG78214.1 PCG78213.1 KC469893 AGC92703.2 ODYU01001380 SOQ37468.1 JN175123 AEN68466.1 JN175099 AEN68442.1 JN175086 AEN68429.1 JN175083 AEN68426.1 JN175092 AEN68435.1 JN175084 AEN68427.1 JN175101 AEN68444.1 JN175104 AEN68447.1 JN175117 AEN68460.1 JN175085 JN175093 AEN68428.1 AEN68436.1 JN175113 AEN68456.1 JN175100 AEN68443.1 JN175082 AEN68425.1 JN175122 AEN68465.1 JN175112 AEN68455.1 JN175079 AEN68422.1 JN175094 AEN68437.1 JN175106 JN175118 JN175119 AEN68449.1 AEN68461.1 AEN68462.1 JN175109 AEN68452.1 JN175089 AEN68432.1 JN175116 AEN68459.1 JN175103 AEN68446.1 JN175115 AEN68458.1 JN175120 AEN68463.1 JN175111 AEN68454.1 JN175097 AEN68440.1 JN175096 AEN68439.1 JN175102 AEN68445.1 JN175081 AEN68424.1 JN175110 JN175121 AEN68453.1 AEN68464.1 JN175078 AEN68421.1 JN175105 JN175107 JN175108 AEN68448.1 AEN68450.1 AEN68451.1 JN175080 AEN68423.1 JN175088 AEN68431.1 JN175114 AEN68457.1 JN175091 AEN68434.1 JN175098 AEN68441.1 JN175090 AEN68433.1 JN175087 AEN68430.1 JN175095 AEN68438.1 AJVK01033236 AJVK01033237 DS235093 EEB11835.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF82708
SSF82708
Gene 3D
ProteinModelPortal
H9J7Z2
A0A194QXJ0
A0A194QGR9
A0A3S2LJM5
A0A1E1WDV1
A0A212EP74
+ More
D0AB98 A0A2A4K2T6 A0A2A4K355 L7X1P9 A0A2H1V9F9 G3KAV1 G3KAS7 G3KAR4 G3KAR1 G3KAS0 G3KAR2 G3KAS9 G3KAT2 G3KAU5 G3KAS1 G3KAU1 G3KAS8 G3KAR0 G3KAV0 G3KAU0 G3KAQ7 G3KAS2 G3KAU7 G3KAT7 G3KAR7 G3KAU4 G3KAT1 G3KAU3 G3KAU8 G3KAT9 G3KAS5 G3KAS4 G3KAT0 G3KAQ9 G3KAT8 G3KAQ6 G3KAT5 G3KAQ8 G3KAR6 G3KAU2 G3KAR9 G3KAS6 G3KAR8 G3KAR5 G3KAS3 A0A1B0GP93 E0VEM9
D0AB98 A0A2A4K2T6 A0A2A4K355 L7X1P9 A0A2H1V9F9 G3KAV1 G3KAS7 G3KAR4 G3KAR1 G3KAS0 G3KAR2 G3KAS9 G3KAT2 G3KAU5 G3KAS1 G3KAU1 G3KAS8 G3KAR0 G3KAV0 G3KAU0 G3KAQ7 G3KAS2 G3KAU7 G3KAT7 G3KAR7 G3KAU4 G3KAT1 G3KAU3 G3KAU8 G3KAT9 G3KAS5 G3KAS4 G3KAT0 G3KAQ9 G3KAT8 G3KAQ6 G3KAT5 G3KAQ8 G3KAR6 G3KAU2 G3KAR9 G3KAS6 G3KAR8 G3KAR5 G3KAS3 A0A1B0GP93 E0VEM9
Ontologies
PANTHER
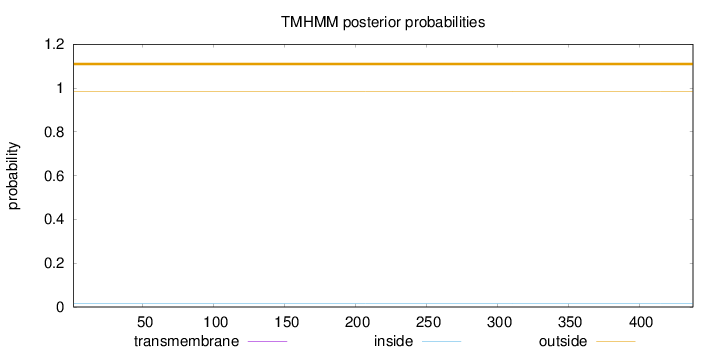
Topology
Length:
438
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01466
outside
1 - 438
Population Genetic Test Statistics
Pi
13.525888
Theta
26.996176
Tajima's D
-1.649329
CLR
0
CSRT
0.0431978401079946
Interpretation
Uncertain
