Gene
KWMTBOMO16489
Pre Gene Modal
BGIBMGA005568
Annotation
putative_NADH-ubiquinone_oxidoreductase_75_kDa_subunit?_mitochondrial_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.17 PlasmaMembrane Reliability : 1.766
Sequence
CDS
ATGTTTAGATTTGCGAGAGAATCATTAACGAAGTTCCAGAAAAATCTTCAAAATGTTGTTAATAGGAGACAGTCAACGCAAGAAATTGATATATTCATTAATGGAAAACCCGTTAAGGTGCCCAGCTACTTCACAATATTACAAGCTTTACGTCGCGAGAAATTGACTGTGCCCACTTTTTGTTATCACGAACGTCTCTCCATTGCTGGTAACTGTAGAATGTGTTTGGTCGAATTAGAAGGCCTGTCAAAGTCACAGGTAGCTTGCGCCATGCCCGTGTCGAAGAACATGAAAATACGAACAAACTCCGCTAAAACCCAAACGGCTCAAGAAGGTGTACTAGAATTTCTACTTGTTAATCATCCATTAGATTGTCCTATTTGTGACCAAGGAGGTGAATGTGATTTGCAAGATCTATCGATGAGATTCGGCAATGACCGTTCAAGGTTTACTGACGTACATTTCGACGGAAAAAGAGCAGTCGAAGATAAAAATTTAGGGCCCTTAATACGAACAGAAATGACTCGTTGCATTCACTGTACTAGATGCATCAGGTTCGCTTCACAAGTTTGTGGCTTGGATGTTTTAGGTACTGCGGGGCGAGGCACTGAAATGCTTGTTGGTACTTATGTCGATAAGATGTTTTTATCTGAGTTATCTGGAAACATCATTGACTTGTGTCCTGTGGGAGCTTTAACATCGATGCCTTACATGTTCAAAGCAAGACCTTGGGAAGTACGGAGAACCAATTCCATTGACGTTACAGATGCAACCGGTACTAATATTGTAATAAATCATAGATTTGATCGTGTTTTAAGGATACTCCCTCGGGAGCACGATGAAATAAACCAAGAATGGCTTTCCGATAAAGGACGGTGGGCCATGGATTCACTTGAAATACAAAGATTAGTGACCCCTATGCTCAGAGTTGGTTCTTGTTTATGTCCTATCGAATGGAATTGTGCGTTGAAAACCGTTGCAGACTCGATAAATTGTACTTGTCCAGAAAATGTAATGGCTATTGCCGGTCCCCATTGTAATGTTGAAACTTTGGTAGCGGCCAAAGACTTTTTAAATATGTTAGGTTCAGAAAACACGTACGTCGAGCGCGGGATGTATTATACACAAACGGGAGCTGATATAAGAGCGGCGTATTCTTTAAATGTAAACATGAGGGATATAGCTCAAGCCGATATAATTTTACTAGTTGGAACCAACCCGAGATTCGAGGCTCCTATTTTGAATGCATGGATCCGACAAGCCTATATTACAAACGAGTGCGATATTTATGTAATAGGTCCTCGATGCGAGTTCAATTATTTTGTCAACTATTTAGGCGATGATATACAATCATTAAGTAAAGCTAGTGAAGTATTAAAAGAAGCTAAAAAACCACTTATTTTAGTTGGAATTTCACAACTGGAATCGGCATTTGGTGGGAAAATCATGAAAGAATTGACTAATATTGCAGCTTCATTTAAGGTAAACAATTGGTCAGTTTTGAATATTGTAACGCGAGAGGCTAGTTTTGCCGGTGCGTTAGAAGCTGGTTGGAAGCCGGGAGCTTTAGCAGCATTACAAGAAAAATGCCCAGATGTTTTGATATCATTGGGTGCTGATGAAATATTTTATAACTGGTCGCCACCACAGTCATGTAAACTTATTTATATAGGTTTTCAAGGTGATAAAGGTTCGGAGAATGCATCTATCGTTTTGCCGGGAAGTGCTTACACCGAAGCCGGTGGATTATATTTGAATATGGAGGGTAGATCTCAACACGCACTTCCTGCAGTCACACCGCCAGGGAAAGCGCGTCACGATTGGAAGATTATACGGGCTTTAGCGGAATATACTGGGATATGTTTGTTCTACAGCGATCACAGTTCTTTATCCTGTCGTCTGTCTCAAGTGAGCCCTAATTTCGTACATTTGGGAAAGTATCAGGATAAATTATTCAGTGACTTGATTGCCCCTCTTATTTGTTCGGATAGCAGTGCGAATGGTAAACTTAATGTCGACATGAAGACTCTGGTTGATTATTATTGTACCGATGTGTTTACTTACAACTCTACGACGATGATAAAGGCCCAGAAAGCTGCAGCAGAGCTTGCGAAATCTAGTTACGCTTGCGTGTAA
Protein
MFRFARESLTKFQKNLQNVVNRRQSTQEIDIFINGKPVKVPSYFTILQALRREKLTVPTFCYHERLSIAGNCRMCLVELEGLSKSQVACAMPVSKNMKIRTNSAKTQTAQEGVLEFLLVNHPLDCPICDQGGECDLQDLSMRFGNDRSRFTDVHFDGKRAVEDKNLGPLIRTEMTRCIHCTRCIRFASQVCGLDVLGTAGRGTEMLVGTYVDKMFLSELSGNIIDLCPVGALTSMPYMFKARPWEVRRTNSIDVTDATGTNIVINHRFDRVLRILPREHDEINQEWLSDKGRWAMDSLEIQRLVTPMLRVGSCLCPIEWNCALKTVADSINCTCPENVMAIAGPHCNVETLVAAKDFLNMLGSENTYVERGMYYTQTGADIRAAYSLNVNMRDIAQADIILLVGTNPRFEAPILNAWIRQAYITNECDIYVIGPRCEFNYFVNYLGDDIQSLSKASEVLKEAKKPLILVGISQLESAFGGKIMKELTNIAASFKVNNWSVLNIVTREASFAGALEAGWKPGALAALQEKCPDVLISLGADEIFYNWSPPQSCKLIYIGFQGDKGSENASIVLPGSAYTEAGGLYLNMEGRSQHALPAVTPPGKARHDWKIIRALAEYTGICLFYSDHSSLSCRLSQVSPNFVHLGKYQDKLFSDLIAPLICSDSSANGKLNVDMKTLVDYYCTDVFTYNSTTMIKAQKAAAELAKSSYACV
Summary
Similarity
Belongs to the complex I 75 kDa subunit family.
Uniprot
H9J7S6
A0A0L7K4G2
A0A3S2TJU3
A0A2A4K1R7
A0A194QB00
A0A194QTG0
+ More
A0A2H1V9G5 A0A212FD95 A0A2H1WGT5 A0A194Q2T0 D6WVN8 E9FYW9 A0A2W1BXT8 A0A0P5BJ82 A0A2A4JWH4 A0A0P6F6M0 A0A0P6AFW5 A0A0P4WX13 A0A0P5JWP1 A0A182SRZ5 A0A0N8AP74 H9JHG0 A0A182WSL6 A0A2C9GQP9 A0A182TLR1 A0A182L9V4 Q7PXY3 A0A2A3E2W2 A0A182PGJ9 A0A0K8TL41 A0A182VBA7 A0A0L7R4U3 J3JZ82 U4USC0 A0A1L8E1A1 A0A182JYV9 A0A1L8E0Q7 A0A0P6G8E9 A0A084WPQ6 E2AUE4 A0A1L8E0U9 A0A226EK77 A0A0K8VPQ5 A0A0V0G757 A0A182VZF8 A0A1A9WX49 A0A0K8USY1 A0A182YB51 A0A182MRG7 A0A034WDL1 A0A1B6EGX1 A0A182RZA0 A0A182JC64 A0A164WP08 A0A0P5XMI3 A0A1Y1NKK2 A0A0J7KS69 A0A2M4A4S5 A0A182N3Z9 A0A1B0GP68 A0A0A1WQ44 A0A195CPY7 A0A0P4W3D4 A0A151JVF0 A0A0T6AUX2 A0A2J7R616 A0A0L8IF50 A0A182FMI2 A0A2M4BF83 A0A151WVB1 T1FN77 A0A151J914 V9IKK8 A0A182QFB8 T1E710 W5JLE0 U5EWU1 Q29FR7 A0A1J1HXL3 A0A0P5G678 W5MY33 A0A195BJI3 U5EIM0 A0A0P5K9E0 A0A0M8ZNA4 E0VE52 A0A224XJ10 A0A0P4WLN3 A0A026VYK1 E9J1P4 A0A154P7K9 A0A1B0GEG7 A0A2M3YZ75 T1HUG6 W8B5N8 A0A1W4X8X9 A0A1W4XJ55 A0A3B0K955 A0A182IMF1 W4YD16
A0A2H1V9G5 A0A212FD95 A0A2H1WGT5 A0A194Q2T0 D6WVN8 E9FYW9 A0A2W1BXT8 A0A0P5BJ82 A0A2A4JWH4 A0A0P6F6M0 A0A0P6AFW5 A0A0P4WX13 A0A0P5JWP1 A0A182SRZ5 A0A0N8AP74 H9JHG0 A0A182WSL6 A0A2C9GQP9 A0A182TLR1 A0A182L9V4 Q7PXY3 A0A2A3E2W2 A0A182PGJ9 A0A0K8TL41 A0A182VBA7 A0A0L7R4U3 J3JZ82 U4USC0 A0A1L8E1A1 A0A182JYV9 A0A1L8E0Q7 A0A0P6G8E9 A0A084WPQ6 E2AUE4 A0A1L8E0U9 A0A226EK77 A0A0K8VPQ5 A0A0V0G757 A0A182VZF8 A0A1A9WX49 A0A0K8USY1 A0A182YB51 A0A182MRG7 A0A034WDL1 A0A1B6EGX1 A0A182RZA0 A0A182JC64 A0A164WP08 A0A0P5XMI3 A0A1Y1NKK2 A0A0J7KS69 A0A2M4A4S5 A0A182N3Z9 A0A1B0GP68 A0A0A1WQ44 A0A195CPY7 A0A0P4W3D4 A0A151JVF0 A0A0T6AUX2 A0A2J7R616 A0A0L8IF50 A0A182FMI2 A0A2M4BF83 A0A151WVB1 T1FN77 A0A151J914 V9IKK8 A0A182QFB8 T1E710 W5JLE0 U5EWU1 Q29FR7 A0A1J1HXL3 A0A0P5G678 W5MY33 A0A195BJI3 U5EIM0 A0A0P5K9E0 A0A0M8ZNA4 E0VE52 A0A224XJ10 A0A0P4WLN3 A0A026VYK1 E9J1P4 A0A154P7K9 A0A1B0GEG7 A0A2M3YZ75 T1HUG6 W8B5N8 A0A1W4X8X9 A0A1W4XJ55 A0A3B0K955 A0A182IMF1 W4YD16
Pubmed
EMBL
BABH01020190
JTDY01010611
KOB58158.1
RSAL01000090
RVE48088.1
NWSH01000227
+ More
PCG78211.1 KQ459232 KPJ02642.1 KQ461155 KPJ08290.1 ODYU01001380 SOQ37469.1 AGBW02009093 OWR51700.1 ODYU01008542 SOQ52237.1 KQ459562 KPI99867.1 KQ971357 EFA09221.1 GL732527 EFX87607.1 KZ149902 PZC78454.1 GDIP01184276 JAJ39126.1 NWSH01000489 PCG76048.1 GDIQ01060543 JAN34194.1 GDIP01029992 JAM73723.1 GDIP01252115 JAI71286.1 GDIQ01197951 JAK53774.1 GDIQ01256467 JAJ95257.1 BABH01012504 BABH01012505 BABH01012506 BABH01012507 APCN01000898 AAAB01008987 EAA00921.4 EGK97518.1 KZ288445 PBC25652.1 GDAI01002544 JAI15059.1 KQ414654 KOC65900.1 APGK01035707 BT128563 KB740928 AEE63520.1 ENN77918.1 KB632428 ERL95413.1 GFDF01001809 JAV12275.1 GFDF01001807 JAV12277.1 GDIQ01037043 JAN57694.1 ATLV01025119 KE525369 KFB52200.1 GL442815 EFN63001.1 GFDF01001808 JAV12276.1 LNIX01000003 OXA58105.1 GDHF01011491 GDHF01001581 JAI40823.1 JAI50733.1 GECL01002957 JAP03167.1 GDHF01022532 GDHF01018974 JAI29782.1 JAI33340.1 AXCM01002120 GAKP01007074 JAC51878.1 GEDC01000123 JAS37175.1 LRGB01001151 KZS13443.1 GDIP01069939 JAM33776.1 GEZM01003715 GEZM01003714 JAV96616.1 LBMM01003841 KMQ93064.1 GGFK01002495 MBW35816.1 AJVK01005738 AJVK01005739 AJVK01005740 AJVK01005741 AJVK01005742 AJVK01005743 AJVK01005744 AJVK01005745 GBXI01013652 JAD00640.1 KQ977513 KYN02174.1 GDKW01000637 JAI55958.1 KQ981701 KYN37487.1 LJIG01022746 KRT78931.1 NEVH01006983 PNF36272.1 KQ415854 KOG00068.1 GGFJ01002563 MBW51704.1 KQ982708 KYQ51794.1 AMQM01005159 KB096830 ESO01146.1 KQ979480 KYN21274.1 JR050215 AEY61202.1 AXCN02002451 GAMD01003432 JAA98158.1 ADMH02000836 ETN64916.1 GANO01001256 JAB58615.1 CH379065 EAL31512.2 CVRI01000020 CRK91262.1 GDIQ01250484 JAK01241.1 AHAT01034460 KQ976453 KYM85333.1 GANO01002580 JAB57291.1 GDIQ01213401 JAK38324.1 KQ435989 KOX67705.1 DS235088 EEB11658.1 GFTR01007854 JAW08572.1 GDRN01062949 JAI65029.1 KK107570 QOIP01000005 EZA48847.1 RLU23102.1 GL767674 EFZ13242.1 KQ434831 KZC07823.1 CCAG010012255 GGFM01000823 MBW21574.1 ACPB03005194 GAMC01017954 JAB88601.1 OUUW01000018 SPP89212.1 AAGJ04030728 AAGJ04030729
PCG78211.1 KQ459232 KPJ02642.1 KQ461155 KPJ08290.1 ODYU01001380 SOQ37469.1 AGBW02009093 OWR51700.1 ODYU01008542 SOQ52237.1 KQ459562 KPI99867.1 KQ971357 EFA09221.1 GL732527 EFX87607.1 KZ149902 PZC78454.1 GDIP01184276 JAJ39126.1 NWSH01000489 PCG76048.1 GDIQ01060543 JAN34194.1 GDIP01029992 JAM73723.1 GDIP01252115 JAI71286.1 GDIQ01197951 JAK53774.1 GDIQ01256467 JAJ95257.1 BABH01012504 BABH01012505 BABH01012506 BABH01012507 APCN01000898 AAAB01008987 EAA00921.4 EGK97518.1 KZ288445 PBC25652.1 GDAI01002544 JAI15059.1 KQ414654 KOC65900.1 APGK01035707 BT128563 KB740928 AEE63520.1 ENN77918.1 KB632428 ERL95413.1 GFDF01001809 JAV12275.1 GFDF01001807 JAV12277.1 GDIQ01037043 JAN57694.1 ATLV01025119 KE525369 KFB52200.1 GL442815 EFN63001.1 GFDF01001808 JAV12276.1 LNIX01000003 OXA58105.1 GDHF01011491 GDHF01001581 JAI40823.1 JAI50733.1 GECL01002957 JAP03167.1 GDHF01022532 GDHF01018974 JAI29782.1 JAI33340.1 AXCM01002120 GAKP01007074 JAC51878.1 GEDC01000123 JAS37175.1 LRGB01001151 KZS13443.1 GDIP01069939 JAM33776.1 GEZM01003715 GEZM01003714 JAV96616.1 LBMM01003841 KMQ93064.1 GGFK01002495 MBW35816.1 AJVK01005738 AJVK01005739 AJVK01005740 AJVK01005741 AJVK01005742 AJVK01005743 AJVK01005744 AJVK01005745 GBXI01013652 JAD00640.1 KQ977513 KYN02174.1 GDKW01000637 JAI55958.1 KQ981701 KYN37487.1 LJIG01022746 KRT78931.1 NEVH01006983 PNF36272.1 KQ415854 KOG00068.1 GGFJ01002563 MBW51704.1 KQ982708 KYQ51794.1 AMQM01005159 KB096830 ESO01146.1 KQ979480 KYN21274.1 JR050215 AEY61202.1 AXCN02002451 GAMD01003432 JAA98158.1 ADMH02000836 ETN64916.1 GANO01001256 JAB58615.1 CH379065 EAL31512.2 CVRI01000020 CRK91262.1 GDIQ01250484 JAK01241.1 AHAT01034460 KQ976453 KYM85333.1 GANO01002580 JAB57291.1 GDIQ01213401 JAK38324.1 KQ435989 KOX67705.1 DS235088 EEB11658.1 GFTR01007854 JAW08572.1 GDRN01062949 JAI65029.1 KK107570 QOIP01000005 EZA48847.1 RLU23102.1 GL767674 EFZ13242.1 KQ434831 KZC07823.1 CCAG010012255 GGFM01000823 MBW21574.1 ACPB03005194 GAMC01017954 JAB88601.1 OUUW01000018 SPP89212.1 AAGJ04030728 AAGJ04030729
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000007151 UP000007266 UP000000305 UP000075901 UP000076407 UP000075840 UP000075902 UP000075882 UP000007062 UP000242457 UP000075885 UP000075903 UP000053825 UP000019118 UP000030742 UP000075881 UP000030765 UP000000311 UP000198287 UP000075920 UP000091820 UP000076408 UP000075883 UP000075900 UP000075880 UP000076858 UP000036403 UP000075884 UP000092462 UP000078542 UP000078541 UP000235965 UP000053454 UP000069272 UP000075809 UP000015101 UP000078492 UP000075886 UP000000673 UP000001819 UP000183832 UP000018468 UP000078540 UP000053105 UP000009046 UP000053097 UP000279307 UP000076502 UP000092444 UP000015103 UP000192223 UP000268350 UP000007110
UP000007151 UP000007266 UP000000305 UP000075901 UP000076407 UP000075840 UP000075902 UP000075882 UP000007062 UP000242457 UP000075885 UP000075903 UP000053825 UP000019118 UP000030742 UP000075881 UP000030765 UP000000311 UP000198287 UP000075920 UP000091820 UP000076408 UP000075883 UP000075900 UP000075880 UP000076858 UP000036403 UP000075884 UP000092462 UP000078542 UP000078541 UP000235965 UP000053454 UP000069272 UP000075809 UP000015101 UP000078492 UP000075886 UP000000673 UP000001819 UP000183832 UP000018468 UP000078540 UP000053105 UP000009046 UP000053097 UP000279307 UP000076502 UP000092444 UP000015103 UP000192223 UP000268350 UP000007110
Pfam
Interpro
IPR036010
2Fe-2S_ferredoxin-like_sf
+ More
IPR001041 2Fe-2S_ferredoxin-type
IPR006656 Mopterin_OxRdtase
IPR010228 NADH_UbQ_OxRdtase_Gsu
IPR000283 NADH_UbQ_OxRdtase_75kDa_su_CS
IPR019574 NADH_UbQ_OxRdtase_Gsu_4Fe4S-bd
IPR006963 Mopterin_OxRdtase_4Fe-4S_dom
IPR015405 NuoG_C
IPR027844 DUF4507
IPR003029 S1_domain
IPR028083 Spt6_acidic_N_dom
IPR028231 Spt6_YqgF
IPR006641 YqgF/RNaseH-like_dom
IPR022967 S1_dom
IPR028088 Spt6_HTH_DNA-bd_dom
IPR035018 Spt6_SH2_C
IPR032706 Spt6_HHH
IPR017072 TF_Spt6
IPR012340 NA-bd_OB-fold
IPR036860 SH2_dom_sf
IPR035420 Spt6_SH2
IPR035019 Spt6_SH2_N
IPR012337 RNaseH-like_sf
IPR000980 SH2
IPR010994 RuvA_2-like
IPR001041 2Fe-2S_ferredoxin-type
IPR006656 Mopterin_OxRdtase
IPR010228 NADH_UbQ_OxRdtase_Gsu
IPR000283 NADH_UbQ_OxRdtase_75kDa_su_CS
IPR019574 NADH_UbQ_OxRdtase_Gsu_4Fe4S-bd
IPR006963 Mopterin_OxRdtase_4Fe-4S_dom
IPR015405 NuoG_C
IPR027844 DUF4507
IPR003029 S1_domain
IPR028083 Spt6_acidic_N_dom
IPR028231 Spt6_YqgF
IPR006641 YqgF/RNaseH-like_dom
IPR022967 S1_dom
IPR028088 Spt6_HTH_DNA-bd_dom
IPR035018 Spt6_SH2_C
IPR032706 Spt6_HHH
IPR017072 TF_Spt6
IPR012340 NA-bd_OB-fold
IPR036860 SH2_dom_sf
IPR035420 Spt6_SH2
IPR035019 Spt6_SH2_N
IPR012337 RNaseH-like_sf
IPR000980 SH2
IPR010994 RuvA_2-like
SUPFAM
Gene 3D
ProteinModelPortal
H9J7S6
A0A0L7K4G2
A0A3S2TJU3
A0A2A4K1R7
A0A194QB00
A0A194QTG0
+ More
A0A2H1V9G5 A0A212FD95 A0A2H1WGT5 A0A194Q2T0 D6WVN8 E9FYW9 A0A2W1BXT8 A0A0P5BJ82 A0A2A4JWH4 A0A0P6F6M0 A0A0P6AFW5 A0A0P4WX13 A0A0P5JWP1 A0A182SRZ5 A0A0N8AP74 H9JHG0 A0A182WSL6 A0A2C9GQP9 A0A182TLR1 A0A182L9V4 Q7PXY3 A0A2A3E2W2 A0A182PGJ9 A0A0K8TL41 A0A182VBA7 A0A0L7R4U3 J3JZ82 U4USC0 A0A1L8E1A1 A0A182JYV9 A0A1L8E0Q7 A0A0P6G8E9 A0A084WPQ6 E2AUE4 A0A1L8E0U9 A0A226EK77 A0A0K8VPQ5 A0A0V0G757 A0A182VZF8 A0A1A9WX49 A0A0K8USY1 A0A182YB51 A0A182MRG7 A0A034WDL1 A0A1B6EGX1 A0A182RZA0 A0A182JC64 A0A164WP08 A0A0P5XMI3 A0A1Y1NKK2 A0A0J7KS69 A0A2M4A4S5 A0A182N3Z9 A0A1B0GP68 A0A0A1WQ44 A0A195CPY7 A0A0P4W3D4 A0A151JVF0 A0A0T6AUX2 A0A2J7R616 A0A0L8IF50 A0A182FMI2 A0A2M4BF83 A0A151WVB1 T1FN77 A0A151J914 V9IKK8 A0A182QFB8 T1E710 W5JLE0 U5EWU1 Q29FR7 A0A1J1HXL3 A0A0P5G678 W5MY33 A0A195BJI3 U5EIM0 A0A0P5K9E0 A0A0M8ZNA4 E0VE52 A0A224XJ10 A0A0P4WLN3 A0A026VYK1 E9J1P4 A0A154P7K9 A0A1B0GEG7 A0A2M3YZ75 T1HUG6 W8B5N8 A0A1W4X8X9 A0A1W4XJ55 A0A3B0K955 A0A182IMF1 W4YD16
A0A2H1V9G5 A0A212FD95 A0A2H1WGT5 A0A194Q2T0 D6WVN8 E9FYW9 A0A2W1BXT8 A0A0P5BJ82 A0A2A4JWH4 A0A0P6F6M0 A0A0P6AFW5 A0A0P4WX13 A0A0P5JWP1 A0A182SRZ5 A0A0N8AP74 H9JHG0 A0A182WSL6 A0A2C9GQP9 A0A182TLR1 A0A182L9V4 Q7PXY3 A0A2A3E2W2 A0A182PGJ9 A0A0K8TL41 A0A182VBA7 A0A0L7R4U3 J3JZ82 U4USC0 A0A1L8E1A1 A0A182JYV9 A0A1L8E0Q7 A0A0P6G8E9 A0A084WPQ6 E2AUE4 A0A1L8E0U9 A0A226EK77 A0A0K8VPQ5 A0A0V0G757 A0A182VZF8 A0A1A9WX49 A0A0K8USY1 A0A182YB51 A0A182MRG7 A0A034WDL1 A0A1B6EGX1 A0A182RZA0 A0A182JC64 A0A164WP08 A0A0P5XMI3 A0A1Y1NKK2 A0A0J7KS69 A0A2M4A4S5 A0A182N3Z9 A0A1B0GP68 A0A0A1WQ44 A0A195CPY7 A0A0P4W3D4 A0A151JVF0 A0A0T6AUX2 A0A2J7R616 A0A0L8IF50 A0A182FMI2 A0A2M4BF83 A0A151WVB1 T1FN77 A0A151J914 V9IKK8 A0A182QFB8 T1E710 W5JLE0 U5EWU1 Q29FR7 A0A1J1HXL3 A0A0P5G678 W5MY33 A0A195BJI3 U5EIM0 A0A0P5K9E0 A0A0M8ZNA4 E0VE52 A0A224XJ10 A0A0P4WLN3 A0A026VYK1 E9J1P4 A0A154P7K9 A0A1B0GEG7 A0A2M3YZ75 T1HUG6 W8B5N8 A0A1W4X8X9 A0A1W4XJ55 A0A3B0K955 A0A182IMF1 W4YD16
PDB
5XTI
E-value=0,
Score=1690
Ontologies
PATHWAY
GO
PANTHER
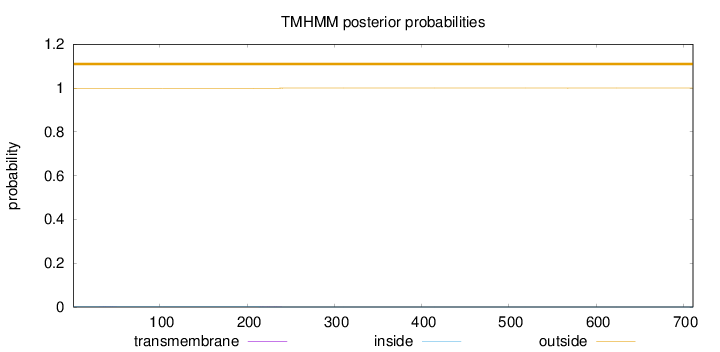
Topology
Length:
711
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0219899999999999
Exp number, first 60 AAs:
0.00311
Total prob of N-in:
0.00109
outside
1 - 711
Population Genetic Test Statistics
Pi
17.573289
Theta
31.67736
Tajima's D
-1.40232
CLR
0.145942
CSRT
0.0711964401779911
Interpretation
Uncertain
