Gene
KWMTBOMO16484 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002222
Annotation
glutathione_S-transferase_delta_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.264 Extracellular Reliability : 1.004
Sequence
CDS
ATGACCATCGACCTGTACTACGTTCCCGGCTCTGCGCCCTGCAGGGCTGTACTGCTGACAGCGAAGGCGCTCAACTTAAACCTGAACCTCAAACTGGTAGACCTACACCACGGGGAACAGCTCAAACCAGAATATTTGAAGTTGAACCCTCAACACACGGTCCCGACCCTAGTCGACGATGGTCTTTCAATCTGGGAGTCCCGCGCCATCATCACCTACTTGGTGAACAAGTACGCCAAAGGCAGCAGTCTGTACCCCGAAGACCCCAAAGCTCGTGCCCTTGTTGACCAACGGTTGTATTTCGACATCGGAACTTTGTACCAGAGATTCAGTGATTATTTCTATCCCCAAGTATTTGCCGGTGCCCCAGCCGACAAGGCCAAGAACGAGAAGGTCCAGGAAGCCCTTCAGCTGCTAGACAAGTTCCTCGAAGGTCAGAAGTACGTGGCCGGACCGAACCTCACCGTCGCCGACCTGAGTCTCATCGCCAGTGTCTCCAGCCTGGAAGCCTCTGACATTGACTTCAAGAAATACGCCAACGTTAAGAGATGGTACGAGACGGTCAAGAGTACAGCTCCTGGATACCAGGAGGCCAACGAGAAAGGACTGGAGGCATTCAAGGGACTTGTCAACAGCATGCTTAAAAAGTGA
Protein
MTIDLYYVPGSAPCRAVLLTAKALNLNLNLKLVDLHHGEQLKPEYLKLNPQHTVPTLVDDGLSIWESRAIITYLVNKYAKGSSLYPEDPKARALVDQRLYFDIGTLYQRFSDYFYPQVFAGAPADKAKNEKVQEALQLLDKFLEGQKYVAGPNLTVADLSLIASVSSLEASDIDFKKYANVKRWYETVKSTAPGYQEANEKGLEAFKGLVNSMLKK
Summary
Similarity
Belongs to the GST superfamily.
Uniprot
Q60GK5
Q964D6
B2CPX9
A0A2S0RQT2
A0A2A4JDN1
B5B2K2
+ More
A0A0K8TD45 A0A212EI48 A0A3G1ZLB2 A0A075X3S8 A0A2H1WG21 A0A0K8TVM2 B0L4I2 S4PAI9 X5CJR6 D7URW9 A0A0K0XRK0 A0A194QFQ3 A0A3S2P3H9 A0A1P8L0S7 Q308N8 A0A194REA6 A0A3G1RJ73 A7KCX1 A0A077D5Y7 J3JZ47 N6UN38 A0A2R4FXG8 U4USZ8 A0A291ARU4 D7NI45 A0A0A7BYH3 A0A2S0BZ20 A0MSN0 A0A3G1RJ77 A0A0A7KQB0 B8YDC1 A0A2A4JDE9 A0A0A7KNK1 X5CCB0 Q5MGM9 A0A0K0XRU3 I4DKS1 A0A194QFP8 A0A2J7QJM2 A0A1P8PET7 I3TBB1 U5EU81 V9I161 A0A139WJ64 T1RTG2 A0A077CXE3 A1XSY6 V5H3B3 A0A0K0XRR2 A0A067QT29 Q2I0J5 A7E1B7 A0A154PQJ5 G8XWV6 A9QUN5 A0A3S2NMH6 A0A161EQE1 I6TU46 A0A2R4NA01 A0A2I6SQK8 A0A1W6L148 U5NM08 A0A1J0F4S9 A0A0K8TV17 A0A1B0FEG6 E9J624 B0YQ88 B0YQ64 B4LXP8 A0A1A9ZZ09 A0A1A9YSW1 G8XWU4 A0A3Q0IV14 T2BBI7 D3TSA7 A0A0G4AJM1 U5NQD8 R4S8C3 A0A194R8U8 U5NLW6 A0A1B0B2P0 B0YQ87 B0YQD1 B0YQ66 B0YQD3 B0YQ63 A0A1A9VMK4 Q5ZFN9 U5NM03 A0A1B6E3T1 R4S4R0 B3M2N3 U5NLM2
A0A0K8TD45 A0A212EI48 A0A3G1ZLB2 A0A075X3S8 A0A2H1WG21 A0A0K8TVM2 B0L4I2 S4PAI9 X5CJR6 D7URW9 A0A0K0XRK0 A0A194QFQ3 A0A3S2P3H9 A0A1P8L0S7 Q308N8 A0A194REA6 A0A3G1RJ73 A7KCX1 A0A077D5Y7 J3JZ47 N6UN38 A0A2R4FXG8 U4USZ8 A0A291ARU4 D7NI45 A0A0A7BYH3 A0A2S0BZ20 A0MSN0 A0A3G1RJ77 A0A0A7KQB0 B8YDC1 A0A2A4JDE9 A0A0A7KNK1 X5CCB0 Q5MGM9 A0A0K0XRU3 I4DKS1 A0A194QFP8 A0A2J7QJM2 A0A1P8PET7 I3TBB1 U5EU81 V9I161 A0A139WJ64 T1RTG2 A0A077CXE3 A1XSY6 V5H3B3 A0A0K0XRR2 A0A067QT29 Q2I0J5 A7E1B7 A0A154PQJ5 G8XWV6 A9QUN5 A0A3S2NMH6 A0A161EQE1 I6TU46 A0A2R4NA01 A0A2I6SQK8 A0A1W6L148 U5NM08 A0A1J0F4S9 A0A0K8TV17 A0A1B0FEG6 E9J624 B0YQ88 B0YQ64 B4LXP8 A0A1A9ZZ09 A0A1A9YSW1 G8XWU4 A0A3Q0IV14 T2BBI7 D3TSA7 A0A0G4AJM1 U5NQD8 R4S8C3 A0A194R8U8 U5NLW6 A0A1B0B2P0 B0YQ87 B0YQD1 B0YQ66 B0YQD3 B0YQ63 A0A1A9VMK4 Q5ZFN9 U5NM03 A0A1B6E3T1 R4S4R0 B3M2N3 U5NLM2
Pubmed
15950511
19121390
22579926
12110299
22118469
30319435
+ More
26017144 17966126 23622113 26198868 26354079 20138238 29912411 17603110 22516182 23537049 28923727 17982444 16023793 22651552 27742358 18362917 19820115 24845553 18716404 24974374 21282665 18245335 17994087 20353571 26252388 23669011 18057021
26017144 17966126 23622113 26198868 26354079 20138238 29912411 17603110 22516182 23537049 28923727 17982444 16023793 22651552 27742358 18362917 19820115 24845553 18716404 24974374 21282665 18245335 17994087 20353571 26252388 23669011 18057021
EMBL
BABH01038371
BABH01038372
BABH01038373
AB176691
BAD60789.1
AF336288
+ More
AAK64362.1 EU541490 ACB36909.1 MF370558 AWA45966.1 NWSH01001884 PCG69798.1 EU887533 FJ360741 ACG69436.1 ACJ23087.1 GBRD01002382 JAG63439.1 AGBW02014704 OWR41162.1 MH177579 AYM01151.1 KF482973 AIH07597.1 ODYU01008429 SOQ52028.1 GCVX01000110 JAI18120.1 EF370473 ABQ53631.1 GAIX01003244 JAA89316.1 KF929201 AHW45899.1 AB541016 BAJ10978.1 KP938858 AKS40339.1 KQ459011 KPJ04373.1 RSAL01000033 RVE51447.1 KX229708 APW77569.1 DQ234273 ABB29466.1 KQ460500 KPJ14261.1 MG879005 AWX68885.1 EF207961 ABS57439.1 KM099179 AIL29309.1 BT128528 AEE63485.1 APGK01025511 KB740594 ENN80137.1 MF034816 AVT42182.1 KB632350 ERL93230.1 KY780632 ATE81124.1 GQ149104 ADD17089.1 KC966933 AHV85207.1 KX255831 ANS53388.1 EF033109 ABK40535.1 MG879006 AWX68886.1 KM433686 AIZ46898.1 FJ517557 FJ517558 ACL51929.1 PCG69796.1 KM433687 AIZ46899.1 KF929202 AHW45900.1 AY829757 AAV91371.1 KP938860 AKS40341.1 AK401889 BAM18511.1 KPJ04368.1 NEVH01013552 PNF28777.1 KU522308 APX61027.1 JQ686954 AFK49803.1 GANO01001603 JAB58268.1 JF793681 AEF98444.1 KQ971338 KYB27875.1 KC569744 AGO46413.1 KJ868730 AIL23531.1 DQ662543 ABG56084.1 GALX01001143 JAB67323.1 KP938859 AKS40340.1 KK853571 KDR06628.1 BABH01038374 BABH01038375 DQ355374 ABC79690.1 AM778448 CAO85744.1 KQ435040 KZC14171.1 FJ855498 AEV23880.1 EU266076 ABX57814.1 RVE51446.1 KR028425 AML23851.1 JN989966 AFM78644.1 MF503496 AVX32554.1 KY549656 AUO28661.1 KX609434 ARN17841.1 KF365938 AGY31900.1 KX018744 APC23398.1 GCVX01000119 JAI18111.1 CCAG010001985 GL768221 EFZ11753.1 EU079405 ABW76219.1 EU079381 EU079385 EU079387 ABW76195.1 ABW76199.1 ABW76201.1 CH940650 EDW67857.1 FJ855485 FJ855486 KY661296 AEV23867.1 AEV23868.1 AVA17383.1 KF061041 AGV08610.1 EZ424309 ADD20585.1 KR133402 AKM70872.1 KF365939 AGY31901.1 KC609530 AGL93467.1 KPJ14263.1 KF365931 AGY31893.1 JXJN01007685 EU079404 EU079409 EU079412 EU079414 EU079415 EU079417 EU079419 EU079423 EU079424 EU079435 EU079438 ABW76218.1 ABW76223.1 ABW76226.1 ABW76228.1 ABW76229.1 ABW76231.1 ABW76233.1 ABW76237.1 ABW76238.1 ABW76249.1 ABW76252.1 EU079448 EU079453 EU079456 ABW76262.1 ABW76267.1 ABW76270.1 EU079383 EU079393 EU079395 EU079396 EU079397 EU079398 EU079399 EU079400 EU079401 EU079402 CH933806 ABW76197.1 ABW76207.1 ABW76209.1 ABW76210.1 ABW76211.1 ABW76212.1 ABW76213.1 ABW76214.1 ABW76215.1 ABW76216.1 EDW14667.1 EU079450 EU079451 EU079452 EU079454 EU079455 EU079457 EU079458 EU079459 EU079460 EU079468 EU079470 ABW76264.1 ABW76265.1 ABW76266.1 ABW76268.1 ABW76269.1 ABW76271.1 ABW76272.1 ABW76273.1 ABW76274.1 ABW76282.1 ABW76284.1 EU079380 EU079382 EU079384 EU079386 EU079388 EU079389 EU079390 EU079391 EU079392 EU079394 ABW76194.1 ABW76196.1 ABW76198.1 ABW76200.1 ABW76202.1 ABW76203.1 ABW76204.1 ABW76205.1 ABW76206.1 ABW76208.1 AJ843575 CAH58743.1 KF365933 AGY31895.1 GEDC01004739 JAS32559.1 KC609534 AGL93471.1 CH902617 EDV42354.1 KF365932 AGY31894.1
AAK64362.1 EU541490 ACB36909.1 MF370558 AWA45966.1 NWSH01001884 PCG69798.1 EU887533 FJ360741 ACG69436.1 ACJ23087.1 GBRD01002382 JAG63439.1 AGBW02014704 OWR41162.1 MH177579 AYM01151.1 KF482973 AIH07597.1 ODYU01008429 SOQ52028.1 GCVX01000110 JAI18120.1 EF370473 ABQ53631.1 GAIX01003244 JAA89316.1 KF929201 AHW45899.1 AB541016 BAJ10978.1 KP938858 AKS40339.1 KQ459011 KPJ04373.1 RSAL01000033 RVE51447.1 KX229708 APW77569.1 DQ234273 ABB29466.1 KQ460500 KPJ14261.1 MG879005 AWX68885.1 EF207961 ABS57439.1 KM099179 AIL29309.1 BT128528 AEE63485.1 APGK01025511 KB740594 ENN80137.1 MF034816 AVT42182.1 KB632350 ERL93230.1 KY780632 ATE81124.1 GQ149104 ADD17089.1 KC966933 AHV85207.1 KX255831 ANS53388.1 EF033109 ABK40535.1 MG879006 AWX68886.1 KM433686 AIZ46898.1 FJ517557 FJ517558 ACL51929.1 PCG69796.1 KM433687 AIZ46899.1 KF929202 AHW45900.1 AY829757 AAV91371.1 KP938860 AKS40341.1 AK401889 BAM18511.1 KPJ04368.1 NEVH01013552 PNF28777.1 KU522308 APX61027.1 JQ686954 AFK49803.1 GANO01001603 JAB58268.1 JF793681 AEF98444.1 KQ971338 KYB27875.1 KC569744 AGO46413.1 KJ868730 AIL23531.1 DQ662543 ABG56084.1 GALX01001143 JAB67323.1 KP938859 AKS40340.1 KK853571 KDR06628.1 BABH01038374 BABH01038375 DQ355374 ABC79690.1 AM778448 CAO85744.1 KQ435040 KZC14171.1 FJ855498 AEV23880.1 EU266076 ABX57814.1 RVE51446.1 KR028425 AML23851.1 JN989966 AFM78644.1 MF503496 AVX32554.1 KY549656 AUO28661.1 KX609434 ARN17841.1 KF365938 AGY31900.1 KX018744 APC23398.1 GCVX01000119 JAI18111.1 CCAG010001985 GL768221 EFZ11753.1 EU079405 ABW76219.1 EU079381 EU079385 EU079387 ABW76195.1 ABW76199.1 ABW76201.1 CH940650 EDW67857.1 FJ855485 FJ855486 KY661296 AEV23867.1 AEV23868.1 AVA17383.1 KF061041 AGV08610.1 EZ424309 ADD20585.1 KR133402 AKM70872.1 KF365939 AGY31901.1 KC609530 AGL93467.1 KPJ14263.1 KF365931 AGY31893.1 JXJN01007685 EU079404 EU079409 EU079412 EU079414 EU079415 EU079417 EU079419 EU079423 EU079424 EU079435 EU079438 ABW76218.1 ABW76223.1 ABW76226.1 ABW76228.1 ABW76229.1 ABW76231.1 ABW76233.1 ABW76237.1 ABW76238.1 ABW76249.1 ABW76252.1 EU079448 EU079453 EU079456 ABW76262.1 ABW76267.1 ABW76270.1 EU079383 EU079393 EU079395 EU079396 EU079397 EU079398 EU079399 EU079400 EU079401 EU079402 CH933806 ABW76197.1 ABW76207.1 ABW76209.1 ABW76210.1 ABW76211.1 ABW76212.1 ABW76213.1 ABW76214.1 ABW76215.1 ABW76216.1 EDW14667.1 EU079450 EU079451 EU079452 EU079454 EU079455 EU079457 EU079458 EU079459 EU079460 EU079468 EU079470 ABW76264.1 ABW76265.1 ABW76266.1 ABW76268.1 ABW76269.1 ABW76271.1 ABW76272.1 ABW76273.1 ABW76274.1 ABW76282.1 ABW76284.1 EU079380 EU079382 EU079384 EU079386 EU079388 EU079389 EU079390 EU079391 EU079392 EU079394 ABW76194.1 ABW76196.1 ABW76198.1 ABW76200.1 ABW76202.1 ABW76203.1 ABW76204.1 ABW76205.1 ABW76206.1 ABW76208.1 AJ843575 CAH58743.1 KF365933 AGY31895.1 GEDC01004739 JAS32559.1 KC609534 AGL93471.1 CH902617 EDV42354.1 KF365932 AGY31894.1
Proteomes
Interpro
ProteinModelPortal
Q60GK5
Q964D6
B2CPX9
A0A2S0RQT2
A0A2A4JDN1
B5B2K2
+ More
A0A0K8TD45 A0A212EI48 A0A3G1ZLB2 A0A075X3S8 A0A2H1WG21 A0A0K8TVM2 B0L4I2 S4PAI9 X5CJR6 D7URW9 A0A0K0XRK0 A0A194QFQ3 A0A3S2P3H9 A0A1P8L0S7 Q308N8 A0A194REA6 A0A3G1RJ73 A7KCX1 A0A077D5Y7 J3JZ47 N6UN38 A0A2R4FXG8 U4USZ8 A0A291ARU4 D7NI45 A0A0A7BYH3 A0A2S0BZ20 A0MSN0 A0A3G1RJ77 A0A0A7KQB0 B8YDC1 A0A2A4JDE9 A0A0A7KNK1 X5CCB0 Q5MGM9 A0A0K0XRU3 I4DKS1 A0A194QFP8 A0A2J7QJM2 A0A1P8PET7 I3TBB1 U5EU81 V9I161 A0A139WJ64 T1RTG2 A0A077CXE3 A1XSY6 V5H3B3 A0A0K0XRR2 A0A067QT29 Q2I0J5 A7E1B7 A0A154PQJ5 G8XWV6 A9QUN5 A0A3S2NMH6 A0A161EQE1 I6TU46 A0A2R4NA01 A0A2I6SQK8 A0A1W6L148 U5NM08 A0A1J0F4S9 A0A0K8TV17 A0A1B0FEG6 E9J624 B0YQ88 B0YQ64 B4LXP8 A0A1A9ZZ09 A0A1A9YSW1 G8XWU4 A0A3Q0IV14 T2BBI7 D3TSA7 A0A0G4AJM1 U5NQD8 R4S8C3 A0A194R8U8 U5NLW6 A0A1B0B2P0 B0YQ87 B0YQD1 B0YQ66 B0YQD3 B0YQ63 A0A1A9VMK4 Q5ZFN9 U5NM03 A0A1B6E3T1 R4S4R0 B3M2N3 U5NLM2
A0A0K8TD45 A0A212EI48 A0A3G1ZLB2 A0A075X3S8 A0A2H1WG21 A0A0K8TVM2 B0L4I2 S4PAI9 X5CJR6 D7URW9 A0A0K0XRK0 A0A194QFQ3 A0A3S2P3H9 A0A1P8L0S7 Q308N8 A0A194REA6 A0A3G1RJ73 A7KCX1 A0A077D5Y7 J3JZ47 N6UN38 A0A2R4FXG8 U4USZ8 A0A291ARU4 D7NI45 A0A0A7BYH3 A0A2S0BZ20 A0MSN0 A0A3G1RJ77 A0A0A7KQB0 B8YDC1 A0A2A4JDE9 A0A0A7KNK1 X5CCB0 Q5MGM9 A0A0K0XRU3 I4DKS1 A0A194QFP8 A0A2J7QJM2 A0A1P8PET7 I3TBB1 U5EU81 V9I161 A0A139WJ64 T1RTG2 A0A077CXE3 A1XSY6 V5H3B3 A0A0K0XRR2 A0A067QT29 Q2I0J5 A7E1B7 A0A154PQJ5 G8XWV6 A9QUN5 A0A3S2NMH6 A0A161EQE1 I6TU46 A0A2R4NA01 A0A2I6SQK8 A0A1W6L148 U5NM08 A0A1J0F4S9 A0A0K8TV17 A0A1B0FEG6 E9J624 B0YQ88 B0YQ64 B4LXP8 A0A1A9ZZ09 A0A1A9YSW1 G8XWU4 A0A3Q0IV14 T2BBI7 D3TSA7 A0A0G4AJM1 U5NQD8 R4S8C3 A0A194R8U8 U5NLW6 A0A1B0B2P0 B0YQ87 B0YQD1 B0YQ66 B0YQD3 B0YQ63 A0A1A9VMK4 Q5ZFN9 U5NM03 A0A1B6E3T1 R4S4R0 B3M2N3 U5NLM2
PDB
3VK9
E-value=5.84538e-105,
Score=970
Ontologies
PATHWAY
00480
Glutathione metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
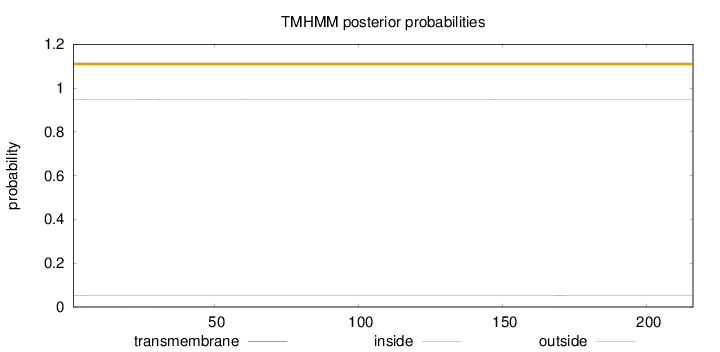
Topology
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0493
Exp number, first 60 AAs:
0.02202
Total prob of N-in:
0.05296
outside
1 - 216
Population Genetic Test Statistics
Pi
22.8811
Theta
32.134746
Tajima's D
-0.75541
CLR
0.010809
CSRT
0.192590370481476
Interpretation
Uncertain
