Gene
KWMTBOMO16473
Pre Gene Modal
BGIBMGA002227
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101736803_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.811
Sequence
CDS
ATGAAGATTCACACCGTTGTGCACAAGATATCACATAATATGACCTACATATTTTCTTTTTGCATTATCTTCGGTACAGTTACGTTTAACCTGACCCCGATTTTCAACAATATCGGGTCGGGTGCATACAAGAATCCTCGACCAGACAACGTGACCTTGCAGCAATGCGTGTATTACGCACTCCCCTTTGACTACACGGGAGATTTCAAGTGGTACTTGCTGGTGGCCATCTTTAACGTACAGAAAACATTTTTCTGCACCTCACTATTTATATTATTTGAATTATCACTGTCCTTGATGATAATATGCTTGTGGGGACATCTCCGGATCTTTATCCACAATTTGAACCACATACCGGCGCCTCGTAACTCCTTGGAGTACACAAAAGAAGAGAGACAAGAAGTGGATGATACGCTGAAGAAGTGCATTCAACACCACACTTTAATAATAGGATTTGTTCGCATAATGTCTGAGACGTACGGTCTGGCGGTCCTCATCTATTACGCTTTCCAGCAAGTGGTTGGATGCTTGTTGCTGTTGCAATGTTCACAAATGGAATTGAAAACTGTCACGCGATTTGGATTTTTGACACTGGTTCTAAATCAACAATTAATTCAAATATCGGTCATATTTGAGCTTTTAGGGTATATGGTAAGTTGCTTTTTTCCCTTTGGAGGTAGATGA
Protein
MKIHTVVHKISHNMTYIFSFCIIFGTVTFNLTPIFNNIGSGAYKNPRPDNVTLQQCVYYALPFDYTGDFKWYLLVAIFNVQKTFFCTSLFILFELSLSLMIICLWGHLRIFIHNLNHIPAPRNSLEYTKEERQEVDDTLKKCIQHHTLIIGFVRIMSETYGLAVLIYYAFQQVVGCLLLLQCSQMELKTVTRFGFLTLVLNQQLIQISVIFELLGYMVSCFFPFGGR
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
Q4W1W4
Q5FBD8
Q4W1W3
D3H5T3
D3H5T2
Q5FBD9
+ More
C4B7U3 B1B1Q0 A7E3F4 B5MEK3 M4SS16 A0A1V1WBX9 A0A2L0NH48 U5KJQ7 A0A1Y9TJH5 A0A1B2G2L9 U5NG03 A0A2H1V437 M9T6E0 T1NXB8 A0A0B4ZYC6 D2KWT5 Q6A1K1 D2KWR9 B5A7F2 A0A2A4KAF9 A0A075T2W9 A0A2A4K9S9 A0A075TD52 C6K7B6 A0A1B3B716 B5MEK0 N0A395 Q6A1J9 C6K7B9 H6VN02 D2KWS9 D2KWQ8 W5ZSF7 B6VGV0 R4JSE8 Q6A1K0 E7FL92 A0A1V1WCF4 D2KWR4 E1ACG1 Q5WA53 Q5WA61 D2KWQ2 A0A1B3B721 Q4W1W6 A0A075T6V4 A0A0C4UU70 A0A345W8A4 A0A2H1W8C8 C7E3V8 U5NJ61 E2S043 A0A1B3B718 A0A2H1W8D7 A0A0E4B3U7 A0A075T6W0 M4NKX7 R4JSE3 D2KWS3 A0A2L0NG62 M9T4N0 A0A2A4K9J7 Q6A1J8 D2KWP6 B5MEJ4 U5KJI4 B5MEK4 E7FL90 T1NXB6 A0A2H1W8J1 A0A0C4UUE4 A0A097IYL9 J7G4H5 Q8MMI0 A0A097ITT1 J7G8K5 A0A2L0NGV3 A0A0S1TQY8 R4JLZ7 A0A075T800 A0A097IYM4 U5KK33 U5KK32 A0A0V0J0Z9 A0A0G2RLW4 Q6A1K3 R4JUU3 A0A0S1TPN2 A0A0B5A1R2 C4MX27 A0A0B5A3B1 C6K7B8 U5NFT5 C6K7C1
C4B7U3 B1B1Q0 A7E3F4 B5MEK3 M4SS16 A0A1V1WBX9 A0A2L0NH48 U5KJQ7 A0A1Y9TJH5 A0A1B2G2L9 U5NG03 A0A2H1V437 M9T6E0 T1NXB8 A0A0B4ZYC6 D2KWT5 Q6A1K1 D2KWR9 B5A7F2 A0A2A4KAF9 A0A075T2W9 A0A2A4K9S9 A0A075TD52 C6K7B6 A0A1B3B716 B5MEK0 N0A395 Q6A1J9 C6K7B9 H6VN02 D2KWS9 D2KWQ8 W5ZSF7 B6VGV0 R4JSE8 Q6A1K0 E7FL92 A0A1V1WCF4 D2KWR4 E1ACG1 Q5WA53 Q5WA61 D2KWQ2 A0A1B3B721 Q4W1W6 A0A075T6V4 A0A0C4UU70 A0A345W8A4 A0A2H1W8C8 C7E3V8 U5NJ61 E2S043 A0A1B3B718 A0A2H1W8D7 A0A0E4B3U7 A0A075T6W0 M4NKX7 R4JSE3 D2KWS3 A0A2L0NG62 M9T4N0 A0A2A4K9J7 Q6A1J8 D2KWP6 B5MEJ4 U5KJI4 B5MEK4 E7FL90 T1NXB6 A0A2H1W8J1 A0A0C4UUE4 A0A097IYL9 J7G4H5 Q8MMI0 A0A097ITT1 J7G8K5 A0A2L0NGV3 A0A0S1TQY8 R4JLZ7 A0A075T800 A0A097IYM4 U5KK33 U5KK32 A0A0V0J0Z9 A0A0G2RLW4 Q6A1K3 R4JUU3 A0A0S1TPN2 A0A0B5A1R2 C4MX27 A0A0B5A3B1 C6K7B8 U5NFT5 C6K7C1
Pubmed
EMBL
AJ874108
CAI43912.1
AB186507
BAD89569.1
AJ874109
CAI43913.1
+ More
FN556592 CBH19583.1 FN556591 CBH19582.1 AB186506 BAD89568.1 AB472092 BAH66309.1 AB186509 BAG12808.1 BK005919 DAA05969.1 AB263205 BAG71423.2 KC299918 AGH58121.1 GENK01000051 JAV45862.1 KY660035 AUY55140.1 KC526967 AGS41443.1 KX084459 ARO76414.1 KU598894 ANZ03140.1 KC960474 AGY14585.2 ODYU01000580 SOQ35608.1 KC188668 AGI96750.1 EU979116 ACL81181.1 KJ542667 AJD81551.1 AB508337 BAI66648.1 AJ748328 CAG38114.1 AB508321 BAI66632.1 EU818705 ACF32964.1 NWSH01000007 PCG80966.1 KF768669 AIG51863.1 PCG80967.1 KF768667 AIG51861.1 GQ165718 ACS45304.1 KT588099 AOE48009.1 AB263116 BAG71420.1 KC538876 AGK43824.1 AJ748330 CAG38116.1 GQ165721 ACS45307.1 JN835269 AEY84942.2 AB508331 BAI66642.1 AB508310 BAI66621.1 KF856285 AHI44516.1 FJ393455 ACJ12370.1 JX982530 AGK90005.1 AJ748329 CAG38115.1 AB597007 BAJ61935.1 GENK01000052 JAV45861.1 AB508316 BAI66627.1 HM595405 LN885099 ADM32897.1 CUQ99388.1 AB101293 BAD69586.1 AB059431 BAD69584.1 AB508304 BAI66615.1 KT588100 AOE48010.1 AJ874106 CAI43910.1 KF768660 AIG51854.1 KJ830840 AIZ94620.1 MF431269 AXJ21642.1 ODYU01006989 SOQ49331.1 GQ246496 ACT34880.1 KC960468 AGY14579.2 AB594450 BAJ22890.1 KT588098 AOE48008.1 SOQ49330.1 LC002701 BAR43449.1 KF768670 AIG51864.1 JX910532 AGG91649.1 JX982525 AGK90000.1 AB508325 BAI66636.1 KY660033 AUY55138.1 KC188669 AGI96751.1 PCG80965.1 AJ748331 CAG38117.1 AB508298 BAI66609.1 AB263110 BAG71414.1 KC526973 AGS41449.1 AB263206 BAG71424.1 AB597005 BAJ61933.1 EU979117 ACL81182.1 SOQ49333.1 KJ830839 AIZ94619.1 KM655531 AIT71981.1 JN701807 AFP66949.1 AJ487481 CAD31948.1 KM597195 AIT69872.1 JN701806 AFP66948.1 KY660036 AUY55141.1 KR935711 ALM26203.1 JX982539 AGK90014.1 KF768703 AIG51897.1 KM655555 AIT72005.1 KC526970 AGS41446.1 KC526965 AGS41441.1 GDKB01000035 JAP38461.1 KJ577702 AJG42377.1 AJ748326 CAG38112.1 JX982531 AGK90006.1 KR935700 ALM26192.1 KJ542669 AJD81553.1 EU791886 ACJ12927.2 KJ542670 AJD81554.1 GQ165720 ACS45306.1 KC960476 AGY14587.2 GQ165723 ACS45309.1
FN556592 CBH19583.1 FN556591 CBH19582.1 AB186506 BAD89568.1 AB472092 BAH66309.1 AB186509 BAG12808.1 BK005919 DAA05969.1 AB263205 BAG71423.2 KC299918 AGH58121.1 GENK01000051 JAV45862.1 KY660035 AUY55140.1 KC526967 AGS41443.1 KX084459 ARO76414.1 KU598894 ANZ03140.1 KC960474 AGY14585.2 ODYU01000580 SOQ35608.1 KC188668 AGI96750.1 EU979116 ACL81181.1 KJ542667 AJD81551.1 AB508337 BAI66648.1 AJ748328 CAG38114.1 AB508321 BAI66632.1 EU818705 ACF32964.1 NWSH01000007 PCG80966.1 KF768669 AIG51863.1 PCG80967.1 KF768667 AIG51861.1 GQ165718 ACS45304.1 KT588099 AOE48009.1 AB263116 BAG71420.1 KC538876 AGK43824.1 AJ748330 CAG38116.1 GQ165721 ACS45307.1 JN835269 AEY84942.2 AB508331 BAI66642.1 AB508310 BAI66621.1 KF856285 AHI44516.1 FJ393455 ACJ12370.1 JX982530 AGK90005.1 AJ748329 CAG38115.1 AB597007 BAJ61935.1 GENK01000052 JAV45861.1 AB508316 BAI66627.1 HM595405 LN885099 ADM32897.1 CUQ99388.1 AB101293 BAD69586.1 AB059431 BAD69584.1 AB508304 BAI66615.1 KT588100 AOE48010.1 AJ874106 CAI43910.1 KF768660 AIG51854.1 KJ830840 AIZ94620.1 MF431269 AXJ21642.1 ODYU01006989 SOQ49331.1 GQ246496 ACT34880.1 KC960468 AGY14579.2 AB594450 BAJ22890.1 KT588098 AOE48008.1 SOQ49330.1 LC002701 BAR43449.1 KF768670 AIG51864.1 JX910532 AGG91649.1 JX982525 AGK90000.1 AB508325 BAI66636.1 KY660033 AUY55138.1 KC188669 AGI96751.1 PCG80965.1 AJ748331 CAG38117.1 AB508298 BAI66609.1 AB263110 BAG71414.1 KC526973 AGS41449.1 AB263206 BAG71424.1 AB597005 BAJ61933.1 EU979117 ACL81182.1 SOQ49333.1 KJ830839 AIZ94619.1 KM655531 AIT71981.1 JN701807 AFP66949.1 AJ487481 CAD31948.1 KM597195 AIT69872.1 JN701806 AFP66948.1 KY660036 AUY55141.1 KR935711 ALM26203.1 JX982539 AGK90014.1 KF768703 AIG51897.1 KM655555 AIT72005.1 KC526970 AGS41446.1 KC526965 AGS41441.1 GDKB01000035 JAP38461.1 KJ577702 AJG42377.1 AJ748326 CAG38112.1 JX982531 AGK90006.1 KR935700 ALM26192.1 KJ542669 AJD81553.1 EU791886 ACJ12927.2 KJ542670 AJD81554.1 GQ165720 ACS45306.1 KC960476 AGY14587.2 GQ165723 ACS45309.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
Q4W1W4
Q5FBD8
Q4W1W3
D3H5T3
D3H5T2
Q5FBD9
+ More
C4B7U3 B1B1Q0 A7E3F4 B5MEK3 M4SS16 A0A1V1WBX9 A0A2L0NH48 U5KJQ7 A0A1Y9TJH5 A0A1B2G2L9 U5NG03 A0A2H1V437 M9T6E0 T1NXB8 A0A0B4ZYC6 D2KWT5 Q6A1K1 D2KWR9 B5A7F2 A0A2A4KAF9 A0A075T2W9 A0A2A4K9S9 A0A075TD52 C6K7B6 A0A1B3B716 B5MEK0 N0A395 Q6A1J9 C6K7B9 H6VN02 D2KWS9 D2KWQ8 W5ZSF7 B6VGV0 R4JSE8 Q6A1K0 E7FL92 A0A1V1WCF4 D2KWR4 E1ACG1 Q5WA53 Q5WA61 D2KWQ2 A0A1B3B721 Q4W1W6 A0A075T6V4 A0A0C4UU70 A0A345W8A4 A0A2H1W8C8 C7E3V8 U5NJ61 E2S043 A0A1B3B718 A0A2H1W8D7 A0A0E4B3U7 A0A075T6W0 M4NKX7 R4JSE3 D2KWS3 A0A2L0NG62 M9T4N0 A0A2A4K9J7 Q6A1J8 D2KWP6 B5MEJ4 U5KJI4 B5MEK4 E7FL90 T1NXB6 A0A2H1W8J1 A0A0C4UUE4 A0A097IYL9 J7G4H5 Q8MMI0 A0A097ITT1 J7G8K5 A0A2L0NGV3 A0A0S1TQY8 R4JLZ7 A0A075T800 A0A097IYM4 U5KK33 U5KK32 A0A0V0J0Z9 A0A0G2RLW4 Q6A1K3 R4JUU3 A0A0S1TPN2 A0A0B5A1R2 C4MX27 A0A0B5A3B1 C6K7B8 U5NFT5 C6K7C1
C4B7U3 B1B1Q0 A7E3F4 B5MEK3 M4SS16 A0A1V1WBX9 A0A2L0NH48 U5KJQ7 A0A1Y9TJH5 A0A1B2G2L9 U5NG03 A0A2H1V437 M9T6E0 T1NXB8 A0A0B4ZYC6 D2KWT5 Q6A1K1 D2KWR9 B5A7F2 A0A2A4KAF9 A0A075T2W9 A0A2A4K9S9 A0A075TD52 C6K7B6 A0A1B3B716 B5MEK0 N0A395 Q6A1J9 C6K7B9 H6VN02 D2KWS9 D2KWQ8 W5ZSF7 B6VGV0 R4JSE8 Q6A1K0 E7FL92 A0A1V1WCF4 D2KWR4 E1ACG1 Q5WA53 Q5WA61 D2KWQ2 A0A1B3B721 Q4W1W6 A0A075T6V4 A0A0C4UU70 A0A345W8A4 A0A2H1W8C8 C7E3V8 U5NJ61 E2S043 A0A1B3B718 A0A2H1W8D7 A0A0E4B3U7 A0A075T6W0 M4NKX7 R4JSE3 D2KWS3 A0A2L0NG62 M9T4N0 A0A2A4K9J7 Q6A1J8 D2KWP6 B5MEJ4 U5KJI4 B5MEK4 E7FL90 T1NXB6 A0A2H1W8J1 A0A0C4UUE4 A0A097IYL9 J7G4H5 Q8MMI0 A0A097ITT1 J7G8K5 A0A2L0NGV3 A0A0S1TQY8 R4JLZ7 A0A075T800 A0A097IYM4 U5KK33 U5KK32 A0A0V0J0Z9 A0A0G2RLW4 Q6A1K3 R4JUU3 A0A0S1TPN2 A0A0B5A1R2 C4MX27 A0A0B5A3B1 C6K7B8 U5NFT5 C6K7C1
Ontologies
GO
PANTHER
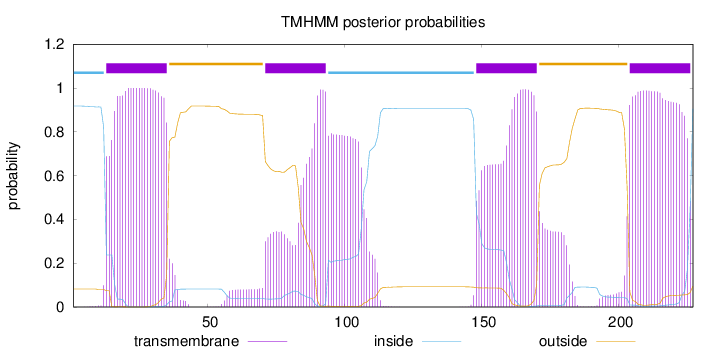
Topology
Subcellular location
Cell membrane
Length:
227
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
92.33175
Exp number, first 60 AAs:
22.82089
Total prob of N-in:
0.91784
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 70
TMhelix
71 - 93
inside
94 - 147
TMhelix
148 - 170
outside
171 - 203
TMhelix
204 - 226
inside
227 - 227
Population Genetic Test Statistics
Pi
15.032747
Theta
22.997578
Tajima's D
-0.977202
CLR
1.525949
CSRT
0.146242687865607
Interpretation
Uncertain
