Gene
KWMTBOMO16470
Pre Gene Modal
BGIBMGA013885
Annotation
PREDICTED:_beta-1?3-galactosyltransferase_4-like?_partial_[Papilio_machaon]
Full name
Hexosyltransferase
Location in the cell
Mitochondrial Reliability : 1.521 PlasmaMembrane Reliability : 1.533
Sequence
CDS
ATGGGCGCGCTGCGACGCACGCACGGCGCGGCGCTGCTGGCGGTCGGGGCCCTCCTACTCGTGCTCTTCGCGCACCTCCCGCCCGCGCCGCGCCCTCCCCCCGCGCCACGACCGGCGCCTCTACTAGTGCCACCCTCCACCCCTCCCGCTCCCTCCAACCGCACAGTTTCCACCCGCGCGTCCCAAACTACAGCGCCGCCGCACCCACCCTCGCCGACCCCGCGACCCAAACCCGCGCCCCTTCTCACTAAGGACGTCTACGTGAGTGGACACGAGCGGCCCCACCCTGAACTGTGTCCGGCGCTGGGGGCACACCTCCGTATGTTAGTGCTGATCACGTCTGCGCCCTCGCACGCTGAGGCGCGCGCTGCCATCCGTTTCACCTGGGGTCACTACGCGGCACGCCGGGACCTCACAATCGCCTTCATGCTGGGCACTCCGCCCCCTGACTTGCGCTCCGCCCTGGTCGCCGAGGACGAGCTGTACGGGGACCTAATTGTGGGCCGTTTCCGAGACTCTTACTCGAACCTCACGCTGAAGACGCTCTCGATGCTGGAGTGGGCGGACACGTACTGCCCGCGAGCGCCGCGCCTCCTGAAGACGGACGACGACATGTTCATCAACGTGCCGCGCCTGCTGCAGTTCGCGGCGGCGCCGGCGCGCGCCAACGCGACCCGCACCATCTGGGGGAAGGTCGTGAAGAAGTCGTACCCGAAGCGCACCACCAAGTCCAAGTACTACGTGTCCCCGCTGCAGTTCTCGGGCAAGGTGTTCCCGGATTTCGCGACGGGGCCGGCCTACCTGCTGACGACCGACGTGGTCCGCGAGCTGCTGGCGGCCGCGCCCCGCGAGCCCTACCTGCGGCTCGAGGACGTGTTCGTGACGGGCGTGCTGGCCGCCAAGCTGCACGTGACGCGCCGCCTCGCGCCGGAGTTCTACAACAAGAAGGTGCAGGCGCACCCCTGCACCGTGCAGCGCGGCGTCGCCATACACATGGTGCGCTACCACGAGCAGTTCGACCTCTGGCGGAAGCTGCTCGACGGCAAGACCAAGTGCGCCAGCTAG
Protein
MGALRRTHGAALLAVGALLLVLFAHLPPAPRPPPAPRPAPLLVPPSTPPAPSNRTVSTRASQTTAPPHPPSPTPRPKPAPLLTKDVYVSGHERPHPELCPALGAHLRMLVLITSAPSHAEARAAIRFTWGHYAARRDLTIAFMLGTPPPDLRSALVAEDELYGDLIVGRFRDSYSNLTLKTLSMLEWADTYCPRAPRLLKTDDDMFINVPRLLQFAAAPARANATRTIWGKVVKKSYPKRTTKSKYYVSPLQFSGKVFPDFATGPAYLLTTDVVRELLAAAPREPYLRLEDVFVTGVLAAKLHVTRRLAPEFYNKKVQAHPCTVQRGVAIHMVRYHEQFDLWRKLLDGKTKCAS
Summary
Similarity
Belongs to the glycosyltransferase 31 family.
Uniprot
A0A2H1VS19
E2AI40
A0A0J7K7Z0
A0A1B6I9I7
A0A1B6LV84
A0A232EJH4
+ More
A0A154PKY5 K7IXW7 A0A195AT65 A0A0L7QLG3 A0A195EL97 E9IGG4 A0A151WJ47 F4W7M5 A0A195ESJ8 V9IGN8 A0A088AUW2 A0A067RKZ6 A0A1W4WZV4 A0A1L8E1E6 A0A1L8E1I3 A0A1L8E1H2 E2C3B0 A0A158NPM8 A0A1B0CM36 A0A151IMP6 A0A0M8ZZJ2 A0A0C9R5P5 A0A2J7RP07 A0A1B0DFY6 A0A026W4H9 D6W731 A0A2R7WUJ0 A0A310SIP0 A0A1S4EJJ1 A0A0A9X6H7 A0A224XSB2 A0A0V0G677 A0A023EY29 A0A2P8ZI62 A0A1J1JB18 A0A1Y1K7E3 A0A1Y1K7D3 A0A0M4EBD2 B4N0Z2 A0A0L0C161 A0A1I8MPA0 T1PD96 A0A1I8NRB9 A0A336M0V7 D6X4Q6 B4LST3 E0VB64 W8AMB2 J3JT67 B4KEC7 A0A1Q3FX53 A0A1Q3FX12 A0A1W4W8D9 A0A0A1XHU6 T1H9L6 B4JC07 B4HYE9 B4H8T3 Q29L44 A0A0K8URA8 A0A182HE01 B4Q6A0 Q9VLS8 A0A3B0K8U2 A0A2M4ALW2 A0A3B0KEM8 A0A1B0AKX6 A0A2M4AL69 B4NWD8 B3N706 A0A034VP52 A0A182J347 A0A1A9W437 A0A023EUN9 B3MMW8 A0A2M3ZKV5 A0A087ZCU3 A0A2M4BNB4 A0A2M4BMR3 A0A2M4BMU1 A0A1A9ZEU0 A0A084VLR0 W5JAY7 A0A182SZY0 A0A1A9XUZ1 A0A182M3N5 A0A1B0FNZ5 A0A1A9UW09 A0A182XBZ9
A0A154PKY5 K7IXW7 A0A195AT65 A0A0L7QLG3 A0A195EL97 E9IGG4 A0A151WJ47 F4W7M5 A0A195ESJ8 V9IGN8 A0A088AUW2 A0A067RKZ6 A0A1W4WZV4 A0A1L8E1E6 A0A1L8E1I3 A0A1L8E1H2 E2C3B0 A0A158NPM8 A0A1B0CM36 A0A151IMP6 A0A0M8ZZJ2 A0A0C9R5P5 A0A2J7RP07 A0A1B0DFY6 A0A026W4H9 D6W731 A0A2R7WUJ0 A0A310SIP0 A0A1S4EJJ1 A0A0A9X6H7 A0A224XSB2 A0A0V0G677 A0A023EY29 A0A2P8ZI62 A0A1J1JB18 A0A1Y1K7E3 A0A1Y1K7D3 A0A0M4EBD2 B4N0Z2 A0A0L0C161 A0A1I8MPA0 T1PD96 A0A1I8NRB9 A0A336M0V7 D6X4Q6 B4LST3 E0VB64 W8AMB2 J3JT67 B4KEC7 A0A1Q3FX53 A0A1Q3FX12 A0A1W4W8D9 A0A0A1XHU6 T1H9L6 B4JC07 B4HYE9 B4H8T3 Q29L44 A0A0K8URA8 A0A182HE01 B4Q6A0 Q9VLS8 A0A3B0K8U2 A0A2M4ALW2 A0A3B0KEM8 A0A1B0AKX6 A0A2M4AL69 B4NWD8 B3N706 A0A034VP52 A0A182J347 A0A1A9W437 A0A023EUN9 B3MMW8 A0A2M3ZKV5 A0A087ZCU3 A0A2M4BNB4 A0A2M4BMR3 A0A2M4BMU1 A0A1A9ZEU0 A0A084VLR0 W5JAY7 A0A182SZY0 A0A1A9XUZ1 A0A182M3N5 A0A1B0FNZ5 A0A1A9UW09 A0A182XBZ9
EC Number
2.4.1.-
Pubmed
20798317
28648823
20075255
21282665
21719571
24845553
+ More
21347285 24508170 30249741 18362917 19820115 25401762 26823975 25474469 29403074 28004739 17994087 26108605 25315136 18057021 20566863 24495485 22516182 23537049 25830018 15632085 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24945155 20920257 24438588
21347285 24508170 30249741 18362917 19820115 25401762 26823975 25474469 29403074 28004739 17994087 26108605 25315136 18057021 20566863 24495485 22516182 23537049 25830018 15632085 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24945155 20920257 24438588
EMBL
ODYU01003804
SOQ43024.1
GL439639
EFN66891.1
LBMM01012025
KMQ86472.1
+ More
GECU01024127 JAS83579.1 GEBQ01012403 JAT27574.1 NNAY01003994 OXU18520.1 KQ434954 KZC12529.1 KQ976745 KYM75386.1 KQ414934 KOC59351.1 KQ978730 KYN28993.1 GL763018 EFZ20339.1 KQ983049 KYQ47854.1 GL887844 EGI69898.1 KQ981986 KYN31198.1 JR044048 AEY59837.1 LC017705 BAR92726.1 KK852564 KDR21275.1 GFDF01001536 JAV12548.1 GFDF01001535 JAV12549.1 GFDF01001537 JAV12547.1 GL452318 EFN77556.1 ADTU01022615 AJWK01018112 AJWK01018113 AJWK01018114 AJWK01018115 KQ977026 KYN06216.1 KQ435794 KOX74117.1 GBYB01011623 GBYB01011624 JAG81390.1 JAG81391.1 NEVH01002143 PNF42567.1 AJVK01033528 AJVK01033529 KK107488 QOIP01000012 EZA49949.1 RLU16078.1 KQ971307 EFA11499.2 KK855596 PTY23218.1 KQ759866 OAD62491.1 GBHO01030934 GDHC01014030 JAG12670.1 JAQ04599.1 GFTR01005557 JAW10869.1 GECL01002537 JAP03587.1 GBBI01004714 JAC13998.1 PYGN01000049 PSN56197.1 CVRI01000074 CRL08241.1 GEZM01090310 JAV57294.1 GEZM01090311 JAV57293.1 CP012523 ALC38722.1 CH963920 EDW78154.2 JRES01001042 KNC26025.1 KA646731 AFP61360.1 UFQT01000375 SSX23690.1 KQ971380 EEZ97677.1 CH940649 EDW64844.1 KRF81894.1 KRF81895.1 DS235022 EEB10620.1 GAMC01019468 JAB87087.1 APGK01047210 BT126423 KB741077 KB631792 AEE61387.1 ENN74230.1 ERL86142.1 CH933807 EDW12895.1 KRG03476.1 KRG03477.1 KRG03478.1 GFDL01002888 JAV32157.1 GFDL01002895 JAV32150.1 GBXI01004214 JAD10078.1 ACPB03024970 CH916368 EDW04110.1 CH480818 EDW52079.1 CH479225 EDW35142.1 CH379061 EAL32980.2 GDHF01023434 JAI28880.1 JXUM01130687 KQ567608 KXJ69346.1 CM000361 CM002910 EDX04176.1 KMY88932.1 AE014134 AY058605 AAF52606.2 AAL13834.1 OUUW01000006 SPP82046.1 GGFK01008277 MBW41598.1 SPP82048.1 JXJN01029118 GGFK01008212 MBW41533.1 CM000157 EDW88455.1 CH954177 EDV59302.1 GAKP01014738 JAC44214.1 GAPW01001424 JAC12174.1 CH902620 EDV30993.1 GGFM01008381 MBW29132.1 GGFJ01005361 MBW54502.1 GGFJ01005235 MBW54376.1 GGFJ01005234 MBW54375.1 ATLV01014545 KE524974 KFB38904.1 ADMH02001755 ETN61166.1 AXCM01000631 CCAG010013125
GECU01024127 JAS83579.1 GEBQ01012403 JAT27574.1 NNAY01003994 OXU18520.1 KQ434954 KZC12529.1 KQ976745 KYM75386.1 KQ414934 KOC59351.1 KQ978730 KYN28993.1 GL763018 EFZ20339.1 KQ983049 KYQ47854.1 GL887844 EGI69898.1 KQ981986 KYN31198.1 JR044048 AEY59837.1 LC017705 BAR92726.1 KK852564 KDR21275.1 GFDF01001536 JAV12548.1 GFDF01001535 JAV12549.1 GFDF01001537 JAV12547.1 GL452318 EFN77556.1 ADTU01022615 AJWK01018112 AJWK01018113 AJWK01018114 AJWK01018115 KQ977026 KYN06216.1 KQ435794 KOX74117.1 GBYB01011623 GBYB01011624 JAG81390.1 JAG81391.1 NEVH01002143 PNF42567.1 AJVK01033528 AJVK01033529 KK107488 QOIP01000012 EZA49949.1 RLU16078.1 KQ971307 EFA11499.2 KK855596 PTY23218.1 KQ759866 OAD62491.1 GBHO01030934 GDHC01014030 JAG12670.1 JAQ04599.1 GFTR01005557 JAW10869.1 GECL01002537 JAP03587.1 GBBI01004714 JAC13998.1 PYGN01000049 PSN56197.1 CVRI01000074 CRL08241.1 GEZM01090310 JAV57294.1 GEZM01090311 JAV57293.1 CP012523 ALC38722.1 CH963920 EDW78154.2 JRES01001042 KNC26025.1 KA646731 AFP61360.1 UFQT01000375 SSX23690.1 KQ971380 EEZ97677.1 CH940649 EDW64844.1 KRF81894.1 KRF81895.1 DS235022 EEB10620.1 GAMC01019468 JAB87087.1 APGK01047210 BT126423 KB741077 KB631792 AEE61387.1 ENN74230.1 ERL86142.1 CH933807 EDW12895.1 KRG03476.1 KRG03477.1 KRG03478.1 GFDL01002888 JAV32157.1 GFDL01002895 JAV32150.1 GBXI01004214 JAD10078.1 ACPB03024970 CH916368 EDW04110.1 CH480818 EDW52079.1 CH479225 EDW35142.1 CH379061 EAL32980.2 GDHF01023434 JAI28880.1 JXUM01130687 KQ567608 KXJ69346.1 CM000361 CM002910 EDX04176.1 KMY88932.1 AE014134 AY058605 AAF52606.2 AAL13834.1 OUUW01000006 SPP82046.1 GGFK01008277 MBW41598.1 SPP82048.1 JXJN01029118 GGFK01008212 MBW41533.1 CM000157 EDW88455.1 CH954177 EDV59302.1 GAKP01014738 JAC44214.1 GAPW01001424 JAC12174.1 CH902620 EDV30993.1 GGFM01008381 MBW29132.1 GGFJ01005361 MBW54502.1 GGFJ01005235 MBW54376.1 GGFJ01005234 MBW54375.1 ATLV01014545 KE524974 KFB38904.1 ADMH02001755 ETN61166.1 AXCM01000631 CCAG010013125
Proteomes
UP000000311
UP000036403
UP000215335
UP000076502
UP000002358
UP000078540
+ More
UP000053825 UP000078492 UP000075809 UP000007755 UP000078541 UP000005203 UP000027135 UP000192223 UP000008237 UP000005205 UP000092461 UP000078542 UP000053105 UP000235965 UP000092462 UP000053097 UP000279307 UP000007266 UP000079169 UP000245037 UP000183832 UP000092553 UP000007798 UP000037069 UP000095301 UP000095300 UP000008792 UP000009046 UP000019118 UP000030742 UP000009192 UP000192221 UP000015103 UP000001070 UP000001292 UP000008744 UP000001819 UP000069940 UP000249989 UP000000304 UP000000803 UP000268350 UP000092460 UP000002282 UP000008711 UP000075880 UP000091820 UP000007801 UP000092445 UP000030765 UP000000673 UP000075901 UP000092443 UP000075883 UP000092444 UP000078200 UP000076407
UP000053825 UP000078492 UP000075809 UP000007755 UP000078541 UP000005203 UP000027135 UP000192223 UP000008237 UP000005205 UP000092461 UP000078542 UP000053105 UP000235965 UP000092462 UP000053097 UP000279307 UP000007266 UP000079169 UP000245037 UP000183832 UP000092553 UP000007798 UP000037069 UP000095301 UP000095300 UP000008792 UP000009046 UP000019118 UP000030742 UP000009192 UP000192221 UP000015103 UP000001070 UP000001292 UP000008744 UP000001819 UP000069940 UP000249989 UP000000304 UP000000803 UP000268350 UP000092460 UP000002282 UP000008711 UP000075880 UP000091820 UP000007801 UP000092445 UP000030765 UP000000673 UP000075901 UP000092443 UP000075883 UP000092444 UP000078200 UP000076407
PRIDE
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A2H1VS19
E2AI40
A0A0J7K7Z0
A0A1B6I9I7
A0A1B6LV84
A0A232EJH4
+ More
A0A154PKY5 K7IXW7 A0A195AT65 A0A0L7QLG3 A0A195EL97 E9IGG4 A0A151WJ47 F4W7M5 A0A195ESJ8 V9IGN8 A0A088AUW2 A0A067RKZ6 A0A1W4WZV4 A0A1L8E1E6 A0A1L8E1I3 A0A1L8E1H2 E2C3B0 A0A158NPM8 A0A1B0CM36 A0A151IMP6 A0A0M8ZZJ2 A0A0C9R5P5 A0A2J7RP07 A0A1B0DFY6 A0A026W4H9 D6W731 A0A2R7WUJ0 A0A310SIP0 A0A1S4EJJ1 A0A0A9X6H7 A0A224XSB2 A0A0V0G677 A0A023EY29 A0A2P8ZI62 A0A1J1JB18 A0A1Y1K7E3 A0A1Y1K7D3 A0A0M4EBD2 B4N0Z2 A0A0L0C161 A0A1I8MPA0 T1PD96 A0A1I8NRB9 A0A336M0V7 D6X4Q6 B4LST3 E0VB64 W8AMB2 J3JT67 B4KEC7 A0A1Q3FX53 A0A1Q3FX12 A0A1W4W8D9 A0A0A1XHU6 T1H9L6 B4JC07 B4HYE9 B4H8T3 Q29L44 A0A0K8URA8 A0A182HE01 B4Q6A0 Q9VLS8 A0A3B0K8U2 A0A2M4ALW2 A0A3B0KEM8 A0A1B0AKX6 A0A2M4AL69 B4NWD8 B3N706 A0A034VP52 A0A182J347 A0A1A9W437 A0A023EUN9 B3MMW8 A0A2M3ZKV5 A0A087ZCU3 A0A2M4BNB4 A0A2M4BMR3 A0A2M4BMU1 A0A1A9ZEU0 A0A084VLR0 W5JAY7 A0A182SZY0 A0A1A9XUZ1 A0A182M3N5 A0A1B0FNZ5 A0A1A9UW09 A0A182XBZ9
A0A154PKY5 K7IXW7 A0A195AT65 A0A0L7QLG3 A0A195EL97 E9IGG4 A0A151WJ47 F4W7M5 A0A195ESJ8 V9IGN8 A0A088AUW2 A0A067RKZ6 A0A1W4WZV4 A0A1L8E1E6 A0A1L8E1I3 A0A1L8E1H2 E2C3B0 A0A158NPM8 A0A1B0CM36 A0A151IMP6 A0A0M8ZZJ2 A0A0C9R5P5 A0A2J7RP07 A0A1B0DFY6 A0A026W4H9 D6W731 A0A2R7WUJ0 A0A310SIP0 A0A1S4EJJ1 A0A0A9X6H7 A0A224XSB2 A0A0V0G677 A0A023EY29 A0A2P8ZI62 A0A1J1JB18 A0A1Y1K7E3 A0A1Y1K7D3 A0A0M4EBD2 B4N0Z2 A0A0L0C161 A0A1I8MPA0 T1PD96 A0A1I8NRB9 A0A336M0V7 D6X4Q6 B4LST3 E0VB64 W8AMB2 J3JT67 B4KEC7 A0A1Q3FX53 A0A1Q3FX12 A0A1W4W8D9 A0A0A1XHU6 T1H9L6 B4JC07 B4HYE9 B4H8T3 Q29L44 A0A0K8URA8 A0A182HE01 B4Q6A0 Q9VLS8 A0A3B0K8U2 A0A2M4ALW2 A0A3B0KEM8 A0A1B0AKX6 A0A2M4AL69 B4NWD8 B3N706 A0A034VP52 A0A182J347 A0A1A9W437 A0A023EUN9 B3MMW8 A0A2M3ZKV5 A0A087ZCU3 A0A2M4BNB4 A0A2M4BMR3 A0A2M4BMU1 A0A1A9ZEU0 A0A084VLR0 W5JAY7 A0A182SZY0 A0A1A9XUZ1 A0A182M3N5 A0A1B0FNZ5 A0A1A9UW09 A0A182XBZ9
Ontologies
GO
PANTHER
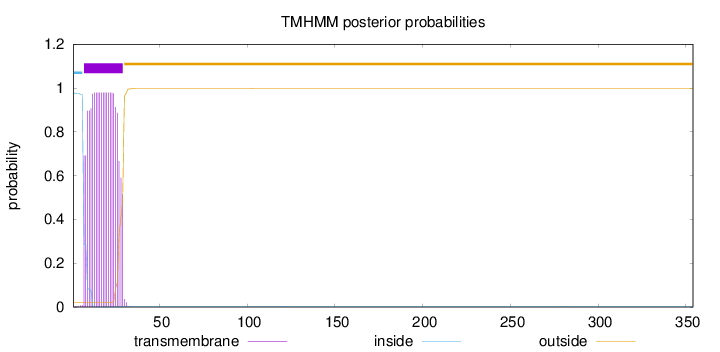
Topology
Subcellular location
Golgi apparatus membrane
SignalP
Position: 1 - 24,
Likelihood: 0.580393
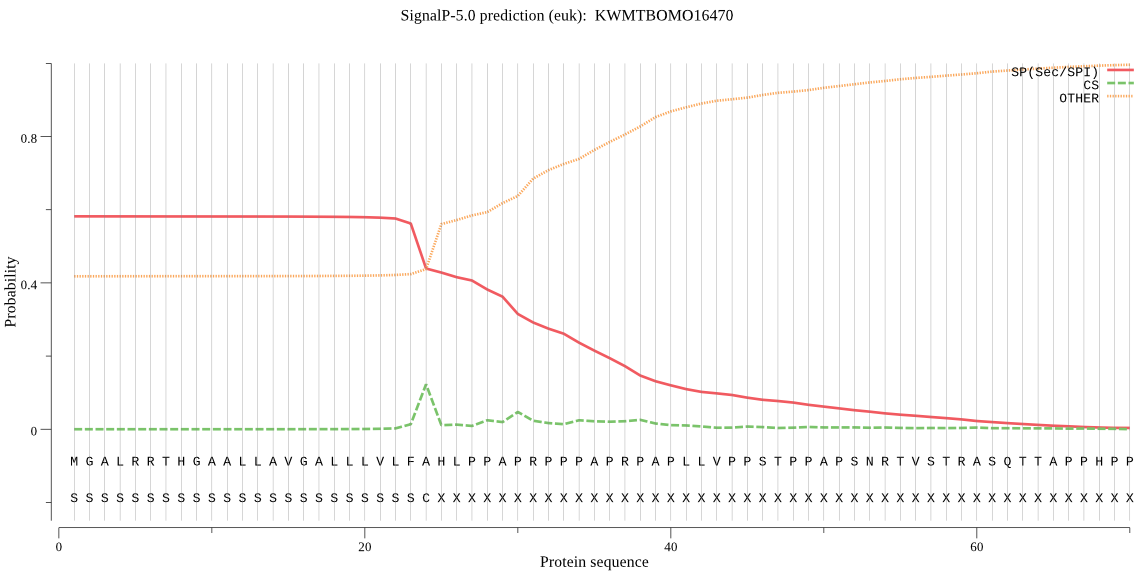
Length:
354
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.4719099999999
Exp number, first 60 AAs:
20.46348
Total prob of N-in:
0.97847
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 354
Population Genetic Test Statistics
Pi
269.457552
Theta
201.135778
Tajima's D
0.966834
CLR
0.296935
CSRT
0.648167591620419
Interpretation
Uncertain
